Research - (2021) Volume 12, Issue 12
Atomic Resolution Snapshot of Eukaryotic 18S Ribosomal RNA of Neglected Tropical Disease (NTDs) Parasites as a Therapeutic Target
Mwangi Harrison Ndung’u1*, Edward Kirwa Muge1, Peter Waiganjo Wagacha2, Albert Ndakala3 and Francis Jackim Mulaa1Abstract
Most of the tropical neglected diseases casing agents are single celled eukaryotic parasites that affect millions of global populations physiologically, economically and sociologically. These diseases not only affect humans but also affect the animals that sustain life within these populations completely influencing the societal fabric on sustenance. The current therapies are limited to poorly selected drugs that have a poor efficacy and extreme adverse effects. The current quick growth in sequencing and atomic structure determination of the parasites cellular organelles has accelerated the venture to obtain newer intervenes that would serve to bene fit these populations. The rapid in silico determination of the ribosomal structure, specifically the 18S rRNA has revealed the mechanisms by which some therapeutics induces their effect on the process of translation to the parasites. We show some in silico atomic resolution structures of 18S rRNA of neglected tropical diseases. We further show strategies and scenarios that are exploited to interfere with translation, thus highlighting the 18S rRNA as a primary drug target. The results also highlight unique as well as conserved elements in binding pocket that can serve as hotspots for novel therapeutics development.
Keywords
Neglected Tropical Diseases (NTDs), 18S rRNA, Ribosome in silico structure determination, Tra nslation.
Introduction
This research work performed as a collaborative project with an eventual goal of developing newer drugs that offer treatment of parasitic infections with lesser side effects, less susceptibility to the mechanism of resistance. The novelty of this work is that it brings to bear the state of art RNA and compound modeling methodologies some of which was developed in the laboratory, to yield high-resolution models of biological molecules and macromolecules. In our case, the smaller subunit ribosomal RNA of the selected parasite was modeled to enable perform virtual screening for new compounds or existing drugs that would allow therapeutic switching. Regions of 18S RNA that have demonstrated the functional significance and highest conservation within the ribosome were identified in this study as targets. The essential nucleotide motifs were selected based on a genetic in vivo system from high throughput identification (Denny SK, et al., 2018; Lee K, et al., 1997). From various studies conducted earlier, there are some regions that have been identified to be essential for the facilitation of protein synthesis from the small subunit (Zafferani M and Hargrove AE, 2021; Sijenyi F, et al., 2012). This includes a region that binds the initiation factor, the tRNA and ribosomal proteins located on helix 23 and helix 24 of on E. coli which is essential to protein synthesis (Schluenzen F, et al., 2000; Stark H, et al., 1997). There are several structural differences between human and parasitic 18S rRNA that could be exploited to tailor specific anti-infectives that only target the rRNA of this pathogenic parasites that are classified as a major class of tropical neglected diseases. To realize this goal we split this into two parts one: the determination of the three-dimensional structure of the selected parasites and identification of the important motif which lies adjacent to helix 23 and 24 of the E. coli and other 2 motifs. Importantly as shown from various studies human helix 24 has a base modification (Pseudouridine) which is lacking in the prokaryotic homolog (Brown DK, et al., 2017; Charette M and Gray MW, 2000; Ofengand J and Bakin A, 1997) it is therefore important to see how it appears for the selected parasites. Determining the structural implication of this particular modified residue toward protein synthesis of these eukaryotes by solving the structures of the chosen parasites hence is important. The next part is performing virtual screening for selected lead compounds by performing lead compound-target recognition. These studies will yield results which will be used to refine further the design strategy of the compound libraries.
Identifying interesting structural domains from the 18S RNA of the chosen parastes, such as helix 23 which forms a domain known as the kink-turn motif (Huang L and Lilley DM, 2018; KleinDJ, et al., 2001) is important. Studies have shown that this motif is highly dynamic (Matsumura S, et al., 2003; Shi X, et al., 2016), hence an excellent target for drugs reason being stabilization of one conformer by a target affects the process of protein synthesis (Mowbray CT, et al., 2003). In addition to this no structure that has been solved completely for an isolated kink-turn (KleinDJ, et al., 2001; Shi X, et al., 2016), therefore characterizing the RNA dynamics and ligand recognition in silico and in vitro offers some difficulty but this could be helped by this study. In silico structure prediction was used to determine the RNA dynamic structures for the parasites. These was followed by identification of interesting motifs sites for drug targets, which were virtually screened with compound databases, and finally this motif-compound binding was validated through the in vitro and finally in vivo techniques.
Introduction to the ribosome
The ribosome is a universal organelle that performs the function of protein synthesis. All parasites have ribosomes that do this process of translation and they are highly conserved due to their functional aspect. Ribosomes are composed of primarily a Ribonucleic Acid (RNA) and several proteins complexed together to achieve a functional unit. These components are either indirectly or directly involved in the process of peptide synthesis. Nucleotide monomers combine to form a polymer known as RNA while amino acid monomers combine to form polymers known as proteins. Due to the diverse functions the RNA molecules play, these molecules are central to the biology of the cell. These functions, for example, may include a host of catalytic roles and various aspects of involvement in gene expression. RNA can adopt different three-dimensional structures to concur with their functional diversity while they interact with other molecules such as protein, cofactors or with other RNAs. It has been observed that there is high sequence conservation in many relevant functional motifs, and this evolutionary conservation points to the important roles that these RNAs have performed throughout time. The desire to unravel underlying mechanisms the various roles that RNA(s) play and the RNA functions has sparked immense interest in structural biology. This is especially due to the surge in the availability of complete genomes sequences. The basic primary methods of elucidating the three-dimensional structure of biological molecules and macromolecules are X-Ray crystallography and Nuclear Magnetic Resonance Spectroscopy though there are some concerted efforts from the cryo-electron microscopy. NMR for isolated RNA domains still remains the method of choice for solution structure determination. However due to the size limitation of NMR towards solving larger RNA newer ways have to be invented (Davis et al., 2005). Newer theoretical methods have been developed that have worked to help minimize the time it takes to obtain the three-dimensional structure of more complex RNA (Mwangi HN, 2013; Mwangi HN, et al., 2021). These include both homology and de novo modeling. The work presented in this study is based on the theoretical methods taking to account highly conserved and functionally relevant ribosomal motifs from the small subunit of the selected parasites (Tables 1 and 2).
| Organism(2) | L(3) | RT(4) | RC | Size | Cmp | Acc | Common name | Phylogeny (M) |
|---|---|---|---|---|---|---|---|---|
| Plasmodium falciparum | N | R | 16S | 2090 | 100 | M19172 | Malaria parasite | Cellular organisms |
| Mycobacterium tuberculosis | N | R | 16S | 1537 | 100 | Z83862 | Cellular organisms | |
| Trymanoma brucei | N | R | 16S | 2251 | 100 | M12676 | Kinetoplastids | Cellular organisms |
| Trypanosoma cruzi | N | R | 16S | 2315 | 100 | AF245382 | Kinetoplastids | Cellular organisms |
| Leishmania major | N | R | 16S | 2203 | 100 | AC005806 | Kinetoplastids | Cellular organisms |
Note: L (3) refers to the cell location, which is the nucleus, RT (4) refers to RNA type R=ribosomal RNA (rRNA), RC refers to the RNA Class 16S, Nucleotide size, Cmp means % Complete, Acc means gene bank accession number, common name and the Phylogenetic Classification, m.
Table 1: Five selected parasites Plasmodium falciparum, M tuberculosis, Trymanoma brucei, Trypanosoma cruzi, and Leishmania major sequences selected for this study
| Species | 18SrRNA. Std. Energy | 18SrRNA. Opt. Energy |
|---|---|---|
| Plasmodium falciparum | 23,97,780.11 | -46,366.56 |
| Mycobacterium tuberculosis | 199459.58 | -37,57,278 |
| Trymanoma brucei | 2118859.871 | -79877.95586 |
| Trypanosoma cruzi | 7455258.998 | -91558.00067 |
| Leishmania major | 895477.8943 | -96921.59635 |
Note: Table obtained from results of RNA 123, which helps minimize the energy from a large positive figure to a more acceptable negative figure that is biologically functional.
Table 2: Total 18S rRNA structure energy optimization
Materials and Methods
The atomic level structural crystalline coordinates used in this study are listed from (Mwangi HN, 2021; Mwangi HN, et al., 2017), along with details of their construction. The sequences used for modeling was conducted using through blasting in the gene bank (NCBI) (Coordinators NR, 2016). Further process of verification was required and led us to specialized groups for verification of the 18S rRNA sequences (CRW) site that has a database that shows the completeness of sequences among other analyzed and verified annotation (Cannone JJ, et al., 2002).
Selection of template structures for the parasites rRNA was a rigorous exercise that is described previously (Mwangi HN, et al., 2021).
The approach for identification of parasites ribosomal RNA drug targets was to utilize the RNA homology modeling software to develop a high throughput method to identify ribosome RNA motif-small molecule partners, or “paired” motifs. Namely three-Dimensional Homology Model Screening (3DHMS) described earlier (Sijenyi F, et al., 2012; Mwangi HN, et al., 2017). Analysis of sensitivity and assays of parasites RNA motif were conducted as described previously (Mwangi HN, et al., 2021).
Refinement, evaluation of 18S rRNA (Sijenyi F, et al., 2012), energy minimization on 18S rRNA (Sijenyi F, et al., 2012; Mwangi HN, et al., 2017), and measurements and prediction of binding to 18S rRNA in the reconstituted structure (Mwangi HN, et al., 2021) were carried out as reported previously, with the minor modifications.
Results and Discussion
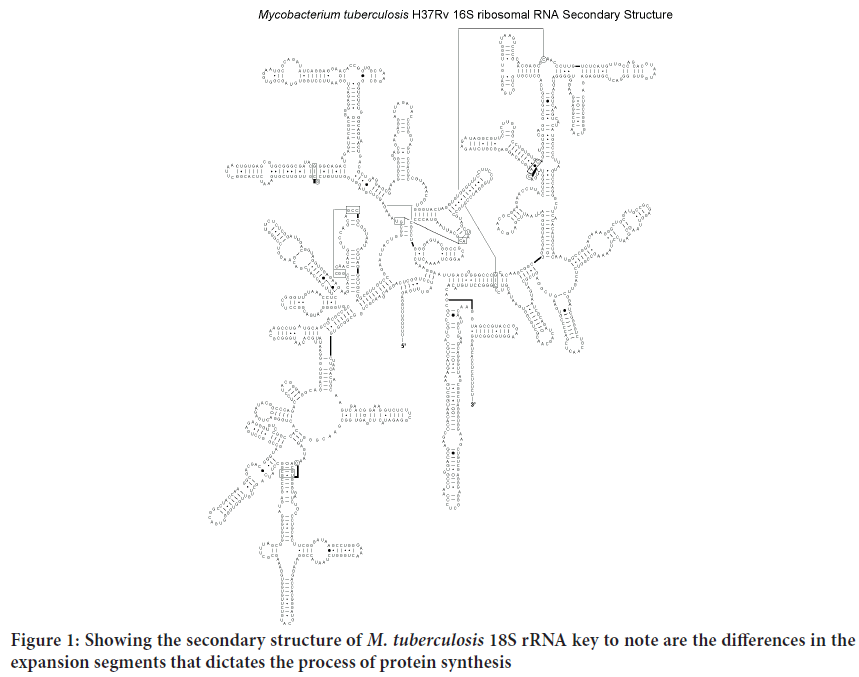
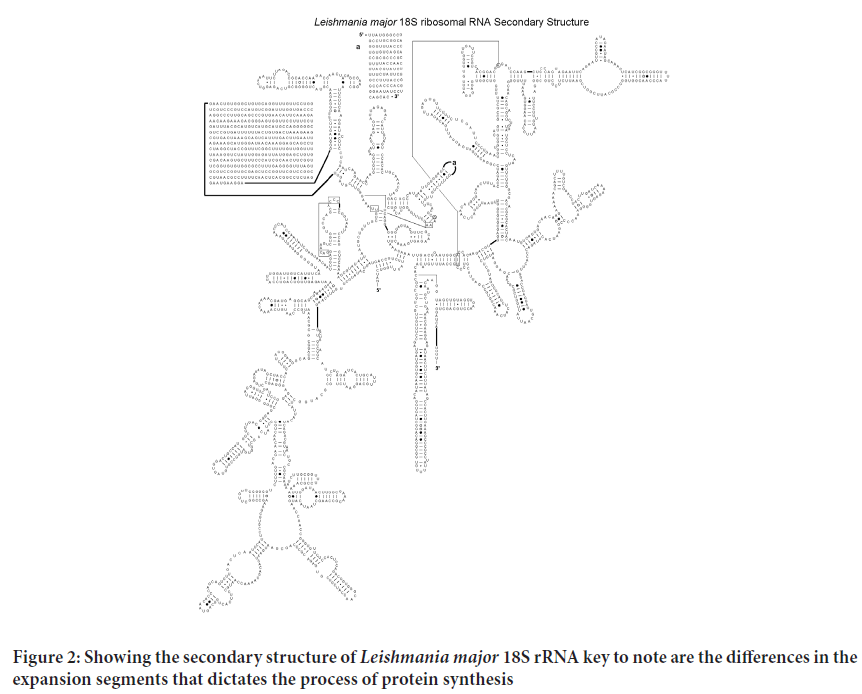
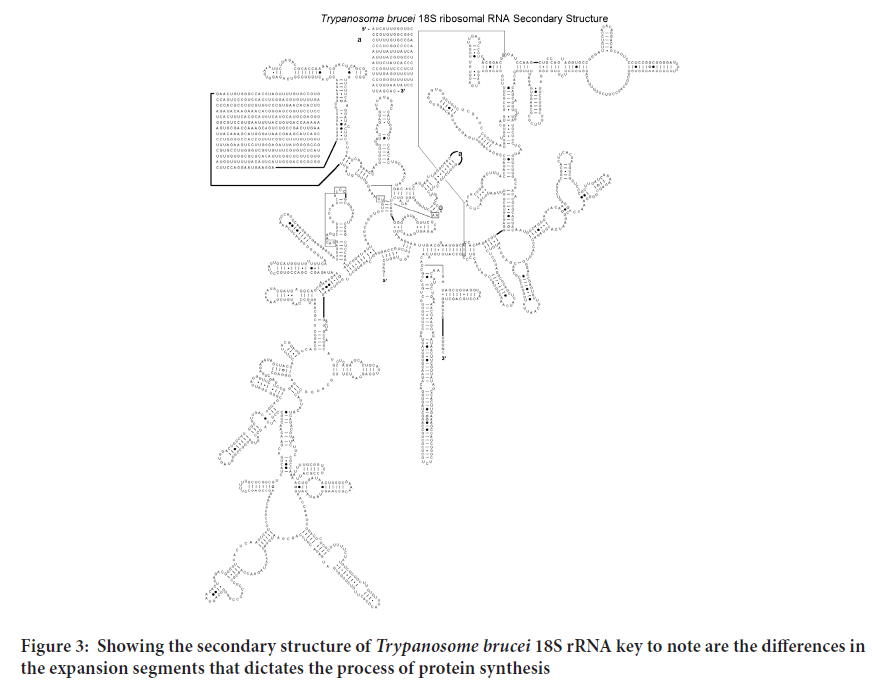
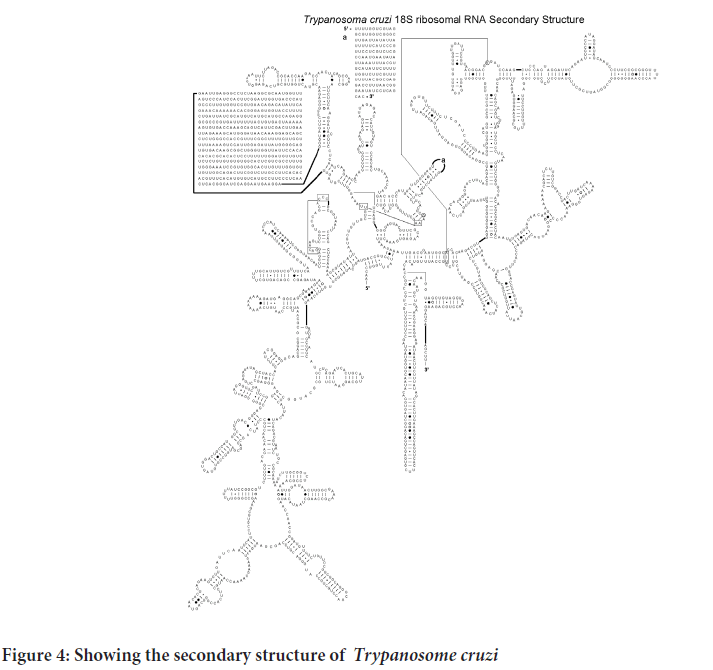
Determining the first structure of the Plasmodium falciparum, Mycobacterium tuberculosis, Trypanosome and Leishmania 18S rRNA has led to better Understanding of the structural basis for its protein-synthesizing roles in these cells. This will enable research in the field of drug development run in silico ligand screening experiments using the solved 18S rRNA structure as a target against a library of potential anti-kinetoplastid compounds. As shown in the Figure 1 to three the various 18S rRNA are highly conserved but have a substantial difference on the expansion segments that lender the process of protein synthesis different from one to the other. The tree dimensional structure further offer differing conformations that could be exploited for potential drug screening and repurposing for more effectiveness towards combating the diseases they cause (Figures 2-4).
Figure 1: Showing the secondary structure of M. tuberculosis 18S rRNA key to note are the differences in the expansion segments that dictates the process of protein synthesis
Figure 2: Showing the secondary structure of Leishmania major 18S rRNA key to note are the differences in the expansion segments that dictates the process of protein synthesis
Figure 3: Showing the secondary structure of Trypanosome brucei 18S rRNA key to note are the differences in the expansion segments that dictates the process of protein synthesis
Figure 4: Showing the secondary structure of Trypanosome cruzi
The use of structure prediction and modeling technologies in this study will dramatically reduce the time it takes from target identification to drug lead determination. Additionally, millions of compounds that can be tested in silico against the generated structure would have been impossible to test experimentally.
Innovation and output
The approaches used in this study to validate and screen rRNA targets are highly innovative. They are superior to traditional screening approaches because of the likelihood of success in finding translation inhibitors that are highly efficacious, but at the same time unlikely to select for target mutations, is very high. Its innovation is based on conserved ribosomal sequences. The exploitation of these sequences could lead to new anti-protozoan drugs refractory to resistance. Drug resistance significant or more likely the highest drug-related priority to date. The production of new anti-protozoan compounds refractory to drug resistance would be a major advance in the repertoire of NTDs. Directing this approach towards other organisms including eukaryotic infective agents could make a significant positive worldwide health impact.
This is an application exploiting the conserved regions of ribosomal rRNA that form functional motifs or loops to develop next-generation anti Plasmodium falciparum, Mycobacterium tuberculosis, Trypanosoma and Leishmania 18S candidate compounds that can be further developed to potent anti Trypanosoma and Leishmania to combat Kinetoplastids family against many protozoans that cause moderate to severe diseases and is applicable to a broad spectrum of apicomplexan protozoan’s diseases.
Conclusion
The application is unique as it proposes to use high-resolution macromolecular models that will lead to the identification of superb modeling systems to the identification of conserved functional motifs of Kinetoplastids ribosomal RNA that for functional reasons cannot be altered and therefore good targets for the screening of lead anti-protozoan compounds.
Acknowledgments
We thank Dr. Fredrick Sijenyi and Dr. Solomon Deresse for insights and comparative reviews on theoretical and experimental techniques to obtain structures, and advice Dr. Khan Nelson for helpful discussion regarding molecular dynamics, Prof Francis Mulaa for valuable advice, and Prof Dimbuson Wallace Bulimo, Miriam Watetu Muratiri and Dr. Daniel Njau for critical comments. This work was supported by the Prof Mulaa Research Program of Nairobi University and by HEP Bioinformatics Consultants LTD.
References
- Denny SK, Bisaria N, Yesselman JD, Das R, Herschlag D, Greenleaf WJ. High-throughput investigation of diverse junction elements in RNA tertiary folding. Cell. 2018; 174(2): 377-390.
- Lee K, Varma S, SantaLucia Jr J, Cunningham PR. In vivo determination of RNA structure-function relationships: Analysis of the 790 loop in ribosomal RNA. J Mol Biol. 1997; 269(5): 732-743.
- Zafferani M, Hargrove AE. Small molecule targeting of biologically relevant RNA tertiary and quaternary structures. Cell Chem Biol. 2021.
- Sijenyi F, Saro P, Ouyang Z, Damm-Ganamet K, Wood M, Jiang J, et al. The RNA folding problems: Different levels of sRNA structure prediction. RNA 3D structure analysis and prediction. 2012; 91-117.
- Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, et al. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell. 2000; 102(5): 615-623.
- Stark H, Orlova EV, Rinke-Appel J, Jünke N, Mueller F, Rodnina M, et al. Arrangement of tRNAs in pre-and post-translocational ribosomes revealed by electron cryomicroscopy. Cell. 1997; 88(1): 19-28.
- Brown DK, Penkler DL, Sheik Amamuddy O, Ross C, Atilgan AR, Atilgan C, et al. MD-TASK: A software suite for analyzing molecular dynamics trajectories. Bioinformatics. 2017; 33(17): 2768-2771.
- Charette M, Gray MW. Pseudouridine in RNA: What, where, how, and why. IUBMB Life. 2000; 49(5): 341-351.
- Ofengand J, Bakin A. Mapping to nucleotide resolution of pseudouridine residues in large subunit ribosomal RNAs from representative eukaryotes, prokaryotes, archaebacteria, mitochondria and chloroplasts. J Mol Biol. 1997; 266(2): 246-268.
- Huang L, Lilley DM. The kink-turn in the structural biology of RNA. Q Rev Biophys. 2018; 51.
- Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink‐turn: A new RNA secondary structure motif. The EMBO Journal. 2001; 20(15): 4214-4221.
- Matsumura S, Ikawa Y, Inoue T. Biochemical characterization of the kink‐turn RNA motif. Nucleic Acids Res. 2003; 31(19): 5544-5551.
- Shi X, Huang L, Lilley DM, Harbury PB, Herschlag D. The solution structural ensembles of RNA kink-turn motifs and their protein complexes. Nat Chem Biol. 2016; 12(3): 146-152.
- Mowbray CT, Holter MC, Teague GB, Bybee D. Fidelity criteria: Development, measurement, and validation. Am J Eval. 2003; 24(3): 315-340.
- Tolbert BS, Miyazaki Y, Barton S, Kinde B, Starck P, Singh R, et al. Major groove width variations in RNA structures determined by NMR and impact of 13 C residual chemical shift anisotropy and 1 H-13 C residual dipolar coupling on refinement. J Biomol NMR. 2010; 47(3): 205-219.
- Mwangi HN. Structure of the 40S ribosomal subunit from Plasmodium falciparum by homology and de novo modeling. Nairobi: University of Nairobi. 2013.
- Mwangi HN, Muge EK, Wagacha PW, Ndakala A, Mulaa FJ. Methods for identifying microbial natural product compounds that target kinetoplastid RNA structural motifs by homology and de novo modeled 18S rRNA. Int J Mol Sci. 2021; 22(9): 4493.
- Mwangi HN, Wagacha P, Mathenge P, Sijenyi F, Mulaa F. Structure of the 40S ribosomal subunit of Plasmodium falciparum by homology and de novo modeling. Acta Pharm Sin B. 2017; 7(1): 97-105.
- Coordinators NR. Database resources of the National Center for Biotechnology Information. 2016.
- Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, et al. The Comparative RNA Web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinform. 2002; 3(1): 1-31.
- Gutell RR, Lee JC, Cannone JJ. The accuracy of ribosomal RNA comparative structure models. Curr Opin Struct. 2002; 12(3): 301-310.
Author Info
Mwangi Harrison Ndung’u1*, Edward Kirwa Muge1, Peter Waiganjo Wagacha2, Albert Ndakala3 and Francis Jackim Mulaa12Department of Computing and Informatics, University of Nairobi, Nairobi, Kenya
3Department of Chemistry, University of Nairobi, Nairobi, Kenya
Received: 21-Jun-2021 Accepted: 05-Jul-2021 Published: 12-Jul-2021
Copyright: This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
ARTICLE TOOLS
- Dental Development between Assisted Reproductive Therapy (Art) and Natural Conceived Children: A Comparative Pilot Study Norzaiti Mohd Kenali, Naimah Hasanah Mohd Fathil, Norbasyirah Bohari, Ahmad Faisal Ismail, Roszaman Ramli SRP. 2020; 11(1): 01-06 » doi: 10.5530/srp.2020.1.01
- Psychometric properties of the World Health Organization Quality of life instrument, short form: Validity in the Vietnamese healthcare context Trung Quang Vo*, Bao Tran Thuy Tran, Ngan Thuy Nguyen, Tram ThiHuyen Nguyen, Thuy Phan Chung Tran SRP. 2020; 11(1): 14-22 » doi: 10.5530/srp.2019.1.3
- A Review of Pharmacoeconomics: the key to “Healthcare for All” Hasamnis AA, Patil SS, Shaik Imam, Narendiran K SRP. 2019; 10(1): s40-s42 » doi: 10.5530/srp.2019.1s.21
- Deuterium Depleted Water as an Adjuvant in Treatment of Cancer Anton Syroeshkin, Olga Levitskaya, Elena Uspenskaya, Tatiana Pleteneva, Daria Romaykina, Daria Ermakova SRP. 2019; 10(1): 112-117 » doi: 10.5530/srp.2019.1.19
- Dental Development between Assisted Reproductive Therapy (Art) and Natural Conceived Children: A Comparative Pilot Study Norzaiti Mohd Kenali, Naimah Hasanah Mohd Fathil, Norbasyirah Bohari, Ahmad Faisal Ismail, Roszaman Ramli SRP. 2020; 11(1): 01-06 » doi: 10.5530/srp.2020.1.01
- Manilkara zapota (L.) Royen Fruit Peel: A Phytochemical and Pharmacological Review Karle Pravin P, Dhawale Shashikant C SRP. 2019; 10(1): 11-14 » doi: 0.5530/srp.2019.1.2
- Pharmacognostic and Phytopharmacological Overview on Bombax ceiba Pankaj Haribhau Chaudhary, Mukund Ganeshrao Tawar SRP. 2019; 10(1): 20-25 » doi: 10.5530/srp.2019.1.4
- A Review of Pharmacoeconomics: the key to “Healthcare for All” Hasamnis AA, Patil SS, Shaik Imam, Narendiran K SRP. 2019; 10(1): s40-s42 » doi: 10.5530/srp.2019.1s.21
- A Prospective Review on Phyto-Pharmacological Aspects of Andrographis paniculata Govindraj Akilandeswari, Arumugam Vijaya Anand, Palanisamy Sampathkumar, Puthamohan Vinayaga Moorthi, Basavaraju Preethi SRP. 2019; 10(1): 15-19 » doi: 10.5530/srp.2019.1.3