Research Article - (2022) Volume 13, Issue 9
Abstract
Nations across the world are currently suffering from various mutations of the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). The ability to infect via respiratory routes such as coughs and sneezes and high mutation rate due to its single strand positive-sense RNA nature makes it very challenging to effectively contain, diagnose, and vaccinate. Moreover, current diagnostic guidelines insist PCR detection for SARS-CoV-2 is done as individual trials for sorting out one mutated variant at a time. To improve the overall diagnosis process and further enhance the efficiency of PCR tests, 4 distinct color emitting probes were used to simultaneously match Delta variant, Omicron variant, positive and negative controls. Amplified signals of individual and merged primers sets were able to successfully detect both Delta and Omicron variants. Additional validation of PCR trials and RNA extracts of SARS-CoV-2 patients further showed the effectiveness of the implemented method. The improved method will further compress the current detection protocol and further benefit diagnosing the ongoing pandemic.
Keywords
Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV-2), Single Nucleotide Polymorphisms (SNP), Delta variant, Omicron variant, Polymerase Chain Reaction (PCR)
Introduction
Among the infamous contagious viruses, Coronavirus has arisen as one of the most concerned public health crises worldwide. Coronaviruses or Coronaviridae are an accumulation of enveloped, positive-strand RNA viruses which not only infects humans, but also a wide selection of amphibians, birds, and mammals. Within the Coronavirus family, two subtypes were already widespread across various nations: Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV-1) and Middle East Respiratory Syndrome-related Coronavirus (MERS-CoV). While both viruses were reported in a worldwide scale, cases and deaths were relatively small compared to other contagious viruses such as the influenza virus (Paget J, et al., 2019), estimating less than 10,000 of both cases and deaths (Feng D, et al., 2009; al Awaidy ST and Khamis F, 2019). However, a subtype derived from SARS-CoV-1 have emerged as a major epidemic later known as SARS-CoV-2 or Coronavirus Disease 2019 (COVID-2019). Unlike its predecessors, SARS-CoV-2 is known to be highly contagious due to its transmission via respiratory droplets from coughs and sneezes which has an infect radius about 1.8 meters (Mittal R, et al., 2020; Jayaweera M, et al., 2020; Bazant MZ and Bush JW, 2021). The ability of transfecting through respiratory methods are not the only advantage. As it is a single strand positive-sense RNA virus, it is able to immediately translate through the host cell DNA compared to negative-sense RNA viruses (Ahlquist P, 2006). Also compared to DNA polymerases, viral RNA polymerases do not have the proofreading ability resulting in a drastically high mutation rate (Prescott LM, et al., 1990; Sanjuán R, et al., 2010). This can arguably be the sole reason why developing effective vaccines against SARS-CoV-2 is a challenge (Sanjuán R, et al., 2010). While most mutations observed in genomes from SARS-CoV-2 are considered either neutral or harmless, mutated regions regarding the spike protein which enables the virus to attach to the surface receptors of host cell membranes are monitored with high attention (Harvey WT, et al., 2021). Not only the spike protein mutation is critical for the virus to successfully dock, but also in the process of linking to host cell receptors, fusion between virus and cell membranes occur which further stimulates the overall mutation of the original strand (Letko M, et al., 2020; Piccoli L, et al., 2020; Liu L, et al., 2020). From the initial outbreak of SARS-CoV-2 virus, 5 major variants are responsible for more than 143 million infections and over three million deaths worldwide (WHO, 2022), B.1.1.7 (Variant Alpha), B.1.351 (Beta variant), P.1 (Gamma variant), B.1.617.2 (Delta variant) and the recently discovered B.1.1.529 (Omicron variant). While the Alpha, Beta and Gamma variants had a severe impact in the global population, the infection and mortality rates were severely decreased thanks to efficient identification vaccination efforts. The more recently occurred Delta and Omicron variants are still a major concern due to mutations of spike proteins such as T478K, P681R L452R, transforming it to a much more contagious state (Starr TN, et al., 2021; Cherian S, et al., 2021). To make matters worse, the Omicron variant is reported to have twice more mutations in gene regions effecting spike proteins (Mannar D, et al., 2022).
The rapid mutations of these recent variants not only hinder making successful vaccines, but also delay the process of identification using methods targeting SNPs and gene mutations. One of the modern procedures of deciding SARS-CoV-2 is by selection of primer sets for specifically targeted genes such as RNA-dependent RNA polymerase (RdRp) (Park M, et al., 2020). However, as more variants, especially Delta and Omicron emerged, the overall selection process has lengthened. Besides the mandatory PCR tests for identifying the common regions such as RdRp, additional amplification of variant specific targets was needed to be individually monitored. This process gradually led to prolonged selection of the overall identification. This study presents an enhanced method of simultaneously targeting Delta and Omicron specific regions while also targeting the common SARS-CoV-2 region RdRp. The compressed method will prove to be more efficient in finding the current rampageous SARS-CoV-2 variants.
Materials and Methods
Primer design
Each primer was designed to match specific point mutations of Delta and Omicron variants. For example, Primer/probe sets targeting the Delta variant were designed to match three distinct variant mutations T19R, L452R and P681R. Likewise, primers targeting the Omicron variant were designed to match three distinct variant mutations T547K, D796Y and Q954H. Within the mutated variant, multiple subtypes indicated the same point mutation among the three regions. Primer/probe sets were then optimized to align in high temperatures to block unwanted non-specific matches. For each targeted point mutation, primers/probes were set at 7 pmol. Positive control RdRp primers and probe were set at 6 pmol.
PCR protocol
Each PCR was performed using the OneStep Multiplex qRT-PCR kit (DiaStar™). The overall platform was designed based on the STexS PCR for accurate mutation detection (Kim JJ, et al., 2021). The master mixture volume was set to 15 μl and the total volume to 20 μl. Reverse transcription was done at 50°C for 2 mins. Initial PCR denaturation was done at 95°C for 5 mins. Denaturation was done at 95°C for 5 secs followed by annealing, extension and detection at 60°C for 3 secs. The process of denaturing to detection was continued for 45 cycles.
SARS-CoV-2 patient samples
RNA extracts from swabbed patients were obtained from the National Culture Collection for Pathogens institute (NCCP) of South Korea (http:// nccp.kdca.go.kr). Sample no. 43408 and no. 43410 were used as standard templates for the Omicron and Delta variant. All samples are publicly accessible and do not require approval from the ethics committee or the institutional review board. All experiments were performed in accordance with the Korean Research Institute of Bioscience and Biotechnology (KRIBB) relevant guidelines and regulations.
Results
Selection of variant specific gene mutations
Apart from the universal SARS-CoV-2 target RdRp, various reports regarding variant specific regions have been identified. Regions distinctly mutated in the Delta variant are the T19R, L452R and P681R region (Cherian S, et al., 2021). The mutation percentage of each gene specific to the variant was estimated around 98.3%, 97.1%, and 99.2% (Table 1). One of the Omicron specifically mutated genes are known to be T547K, D796Y and Q954H (van Blargan LA, et al., 2022). The percentage was also very high ranging from 94% to 98%. These distinct features indicate if primers can pinpoint at least two mutations of any variant, it would designate the patient to be diagnosed as Gamma or Omicron variant SARS-CoV-2.
| Variants of COVID-19 | Target gene (%) | |||||
|---|---|---|---|---|---|---|
| T19R | L452R | P681R | T547K | D796Y | Q954H | |
| Omicron | 0.1 | 0.5 | 0.1 | 93.9 | 96.9 | 97.8 |
| Delta | 98.3 | 97.1 | 99.2 | 0.1 | 0.1 | ND |
| Alpha | ND | 0.1 | 0.1 | 0.1 | 0.1 | ND |
| Beta | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | ND |
| Gamma | 0.1 | 0.1 | 0.4 | 0.1 | 0.1 | ND |
Table 1: Mutation occurrence of target genes among various variants of SARS-CoV-2
Alongside selecting the genes, SNP regions of each target were selected for optimal primer assembling. To enhance the efficiency of the overall SARS-CoV-2 detection process, four distinct colored fluorophores were used to effectively distinguish PCR amplification between SARS-CoV-2, Delta and Omicron variant, and negative control. To decrease false negative results, each fluorophores consisted of probes matching to three point mutations of each variant. To confirm whether two specific regions of each variant could represent a positive match, correlation analysis was done for mutation of three genes T19R, L452R and P681R in Delta variants, T547K, D796Y and Q954H in Omicron variants. Results showed a 99.58% correlation of three variants co-existing in Delta variants, and 99.48% in omicron variants (Table 2). This validates when the two of three mutated regions of each variant were matched, it would indicate a positive diagnose of the patient as Delta or Omicron SARS-CoV-2.
| Omicron variants (10,394) | ||||||||||
| VOC | T547K, D796Y, Q954H | T547K, D796Y | T547K, Q954H | D796Y, Q954H | T547K | D796Y | Q954H | |||
| Omicron | 10,340 (99.48%) | 7 (0.07%) | 27 (0.26%) | 20 (0.19%) | 0 | 0 | 0 | |||
| Delta variants (11,354) | ||||||||||
| VOC | T19R, L452R, P681R | T19R, L452R | T19R, P681R | T547K, P681R | T19R | L452R | P681R | |||
| Delta | 11,306 (99.58%) | 13 (0.11%) | 9 (0.08) | 26 (0.23%) | 0 | 0 | 0 | |||
Table 2: Point mutation correlation of Delta and Omicron variants
Optimization of the Polymerase Chain Reaction (PCR) procedure
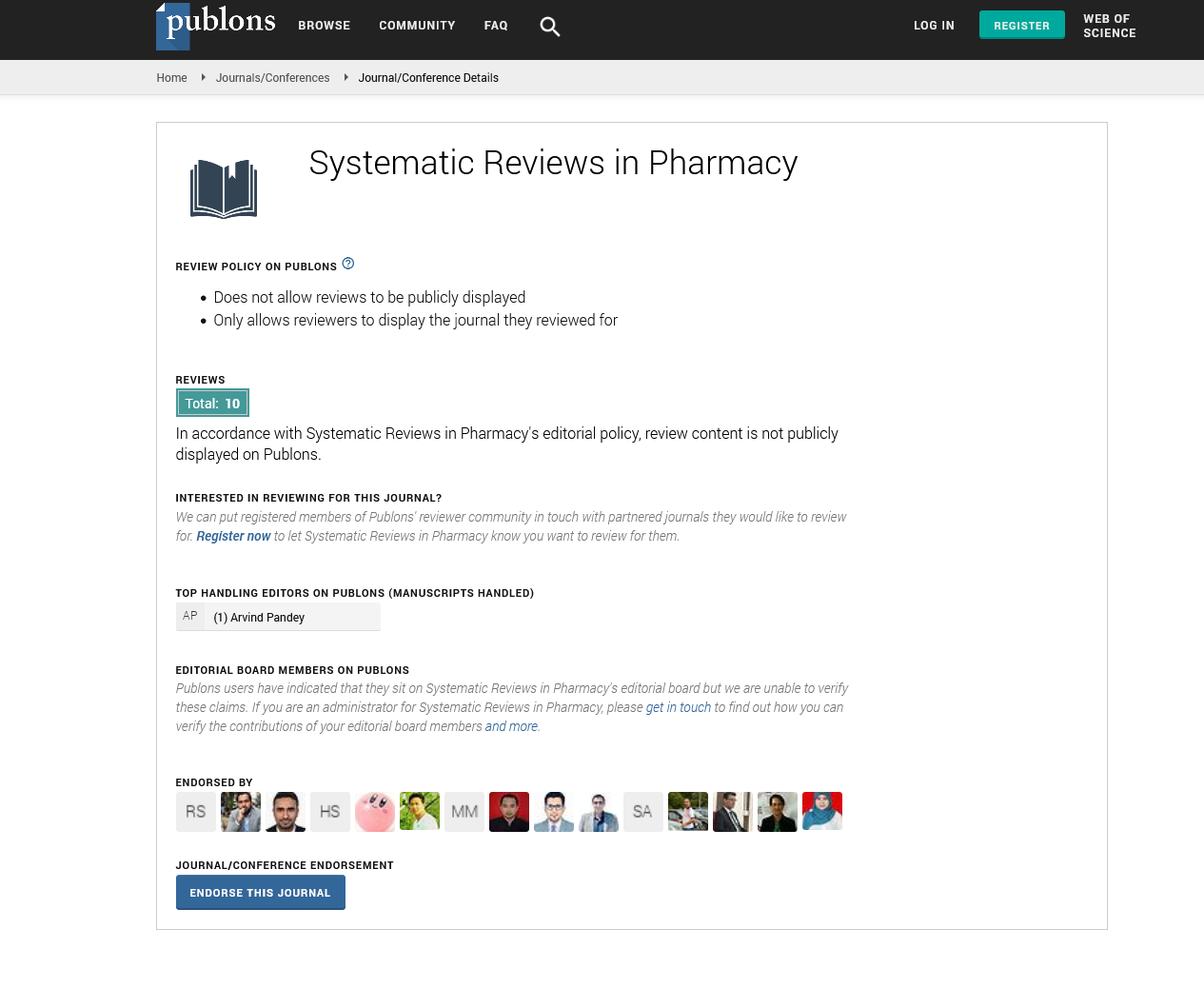
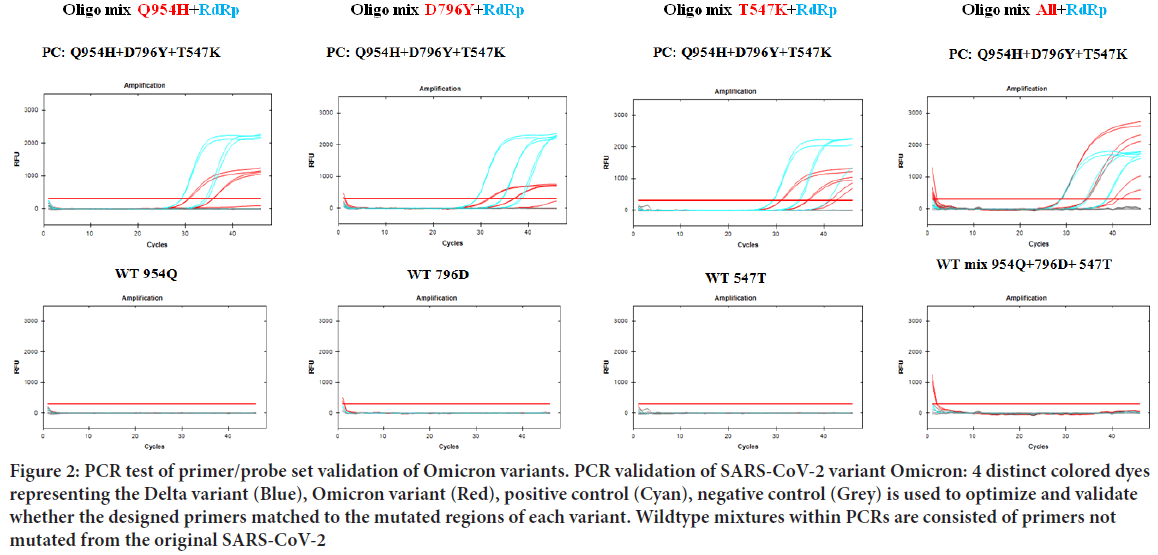
Once the primer targets were set, multiple PCR tests were performed for optimal amplification. All PCR tests were evaluated using standard plasmid DNAs containing inserts corresponding target regions. Amplification of RdRp and Negative control was initially confirmed to rule out possible interference. As suspected, all SARS-CoV-2 variants tested showed high amplified signals of RdRp and no signals of negative controls (Figure 1). PCR trials of Delta and Omicron variants were also performed. For each variant, every primer/probe set was tested with the SARS-CoV-2 positive and negative controls to ensure every amplified signal was detected over the PCR threshold. As a result, amplification signals of T19R, L452R, P681R within the Delta variant and T547K, D796Y, Q954H within the Omicron variant was detected above the PCR threshold (Figure 2). After the validation of each target, three targets altogether of each variant were tested. The detection signal was as strong as the positive control RdRp and was easily distinguishable to be selected as a match. Finally, to confirm the variants were only amplified in a SNP mutated primer, wildtype standard plasmid of each target was tested. As a result, each primer sets for the Delta and Omicron variants did not amplify using the wildtype plasmid DNA as a template, proving no interference of non-specific template annealing.
Figure 1: Polymerase Chain Reaction (PCR) test of primer/probe set validation of Delta variants. PCR validation of SARS-CoV-2 variant Delta: 4 distinct colored dyes representing the Delta variant (Blue), Omicron variant (Red), positive control (Cyan), negative control (Grey) is used to optimize and validate whether the designed primers matched to the mutated regions of each variant. Wildtype mixtures within PCRs are consisted of primers not mutated from the original SARS-CoV-2
Figure 2: PCR test of primer/probe set validation of Omicron variants. PCR validation of SARS-CoV-2 variant Omicron: 4 distinct colored dyes representing the Delta variant (Blue), Omicron variant (Red), positive control (Cyan), negative control (Grey) is used to optimize and validate whether the designed primers matched to the mutated regions of each variant. Wildtype mixtures within PCRs are consisted of primers not mutated from the original SARS-CoV-2
Validation using RNA samples of SARS-CoV-2 patients
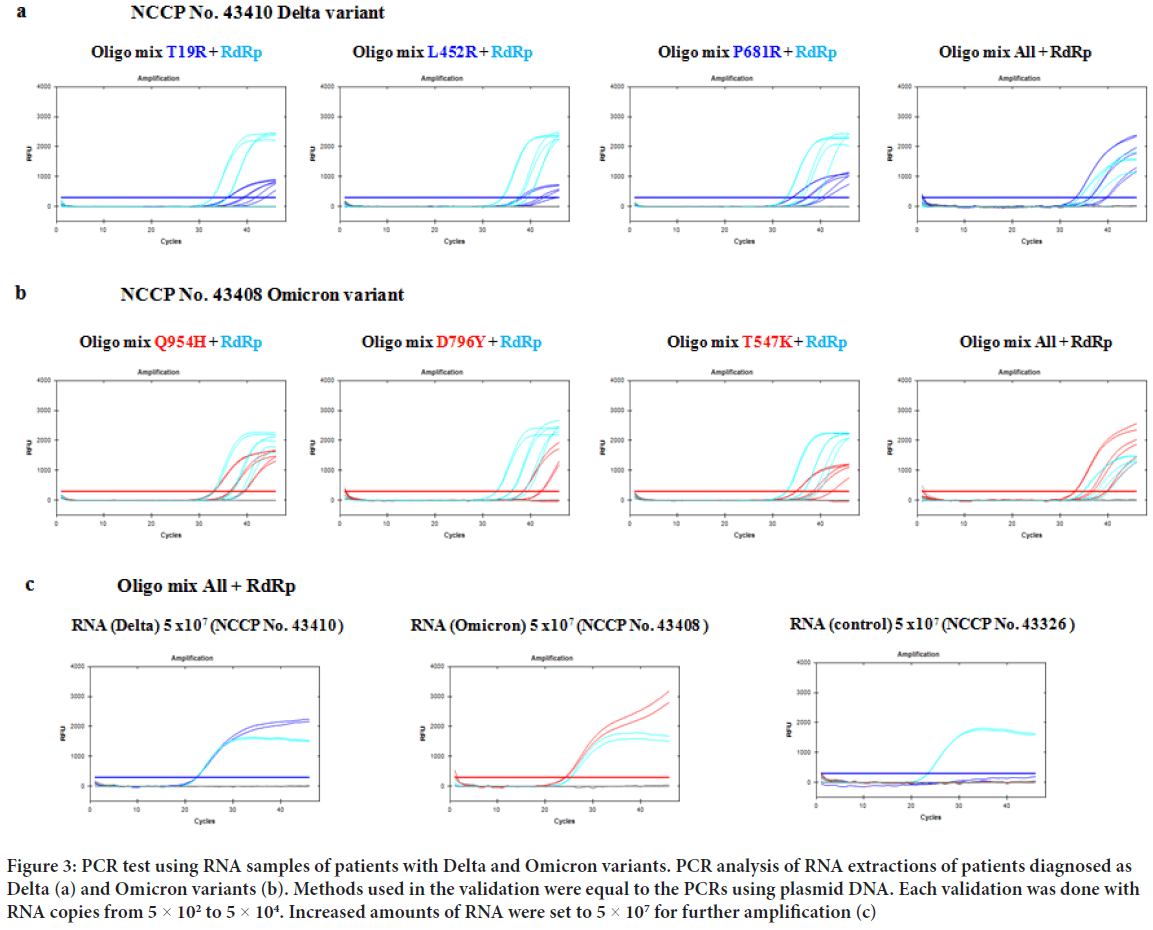
Once the optimization was done, further validation was done using donated RNA extraction of SARS-CoV-2 patients. Each RNA samples were tested with primer/probe targets of T19R, L452R, P681R within the Delta variant and T547K, D796Y, Q954H within the Omicron variant. When comparing the amplified data with the plasmid DNA, individual target signals were in par with the RdRp positive control (Figure 3). When all three primer sets were tested in a single PCR, amplified signals of Delta and Omicron variants were far above the positive control. To further test the amplification specificity, sufficient amount of COVID RNA was included in each PCR validation. When the input was increased to 5 × 107 RNA of both variants, amplified signals surpassed RdRp positive control (Figure 3). The results would mean actual patient PCR trials will be more easily diagnosed.
Figure 3: PCR test using RNA samples of patients with Delta and Omicron variants. PCR analysis of RNA extractions of patients diagnosed as Delta (a) and Omicron variants (b). Methods used in the validation were equal to the PCRs using plasmid DNA. Each validation was done with RNA copies from 5 × 102 to 5 × 104. Increased amounts of RNA were set to 5 × 107 for further amplification (c)
Discussion
The SARS-CoV-2 pandemic is a concurring global public threat, infecting millions and potentially triggering permanent damage in the respiratory system. While research is continuously done to effectively repress, diagnose, and cure the virus, more and more samples of patients potentially infected of SARS-CoV-2 are needed to be immediately tested. Despite of the ever-increasing amount of patient samples and mutated variants, current diagnostic protocols are still required to be performed one variant at a time. This overall time-consuming process can be solved by distinguishing the recent variants simultaneously in one PCR test. The methods mentioned above will serve as an efficient improvement and further benefit on diagnosing Delta and Omicron variants. Furthermore, by confirming positive matches of at least two mutated regions of Delta and Omicron variants, false negative results will be decreased further declining the risk of unsuspected outbreaks.
The concurring mutation of SARS-CoV-2 was followed by variant specific SNPs reported in various institutions. As the research itself is crucial for tackling the imminent treat, the actual patient infected with SARS-CoV-2 will seem irrelevant. The thing that is the most relevant to the individual will be the outcome of the SARS-CoV-2 PCR trial. The method proposed above will be more beneficial to diagnose the Delta and Omicron variants of SARS-CoV-2 due to the combined amplification of each three targets. Further tests on the PCR revealed while individual primer targets were amplified with high sensitivity, merged primers tended to express enhanced specificity as the combined primers amplified with a synergistic manner. The overall process will effectively guide patients and medical personnel to diagnose the current disease.
Conclusion
Despite numerous efforts worldwide in attempts to successfully contain, distinguish, cure, and restore the ever-spreading Coronavirus, it is for certain the termination of the disease will be challenging. As with the previous variations, novel mutations of SARS-CoV-2 may emerge as a major variant, demanding the health care institution additional methods of diagnosis. This will inevitably lengthen the whole process, which will further decelerate the current selection. Combining multiple variant targets in one detection kit will be required to catch up with the SARS-CoV-2 mutation. The suggested method may benefit future studies associating multiple target diagnosis.
Declarations
Author contributions
Jae Jong Kim contributed in the experimental design, PCR, discussions, and project management. Hyoung-Min Park wrote the overall article manuscript and contributed in PCR tests. A Young Kyoung contributed in PCR and primer designs. Si-Kyu Lim contributed in experimental designs, discussions, and article writing. Sun Ho Cha contributed in samples preparation, PCR, and primer preparation. J Eugene Lee contributed in article feedback, discussions, and project management. Byoung Chul Park contributed in article writing, design, and conducting the project. Jae Jong Kim and Hyoung-Min Park both contributed equally to this work.
Acknowledgements
We thank the National Culture Collection for Pathogens institute (NCCP) for providing the SARS-CoV-2 patient RNA samples as a standard template for Delta and Omicron variants. The article has been supported by GENOTEC CORP (Grant number GTR2021-02).
Data availability statement
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
References
- Paget J, Spreeuwenberg P, Charu V, Taylor RJ, Iuliano AD, Bresee J, et al. Global mortality associated with seasonal influenza epidemics: New burden estimates and predictors from the GLaMOR Project. J Glob Health. 2019; 9(2): 20421.
[Crossref] [Google Scholar] [Pubmed]
- Feng D, de Vlas SJ, Fang LQ, Han XN, Zhao WJ, Sheng S, et al. The SARS epidemic in mainland China: Bringing together all epidemiological data. Trop Med Int Health. 2009; 14: 4-13.
[Crossref] [Google Scholar] [Pubmed]
- al Awaidy ST, Khamis F. Middle East Respiratory Syndrome Coronavirus (MERS-CoV) in Oman: Current situation and going forward. Oman Med J. 2019; 34(3): 181.
[Crossref] [Google Scholar] [Pubmed]
- Mittal R, Ni R, Seo JH. The flow physics of COVID-19. J Fluid Mech. 2020; 894.
- Jayaweera M, Perera H, Gunawardana B, Manatunge J. Transmission of COVID-19 virus by droplets and aerosols: A critical review on the unresolved dichotomy. Environ Res. 2020; 188: 109819.
[Crossref] [Google Scholar] [Pubmed]
- Bazant MZ, Bush JW. A guideline to limit indoor airborne transmission of COVID-19. Proc Natl Acad Sci USA. 2021; 118(17): 2018995118.
[Crossref] [Google Scholar] [Pubmed]
- Ahlquist P. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat Rev Microbiol. 2006; 4(5): 371-382.
[Crossref] [Google Scholar] [Pubmed]
- Prescott LM, Harley JP, Klein DA. Microbiology. Wm C Brown Publishers. 1990.
- Sanjuán R, Nebot MR, Chirico N, Mansky LM, Belshaw R. Viral mutation rates. J Virol. 2010; 84(19): 9733-9748.
[Crossref] [Google Scholar] [Pubmed]
- Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021; 19(7): 409-424.
[Crossref] [Google Scholar] [Pubmed]
- Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nature microbiology. 2020; 5(4): 562-569.
[Crossref] [Google Scholar] [Pubmed]
- Piccoli L, Park YJ, Tortorici MA, Czudnochowski N, Walls AC, Beltramello M, et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020; 183(4): 1024-1042.
[Crossref] [Google Scholar] [Pubmed]
- Liu L, Wang P, Nair MS, Yu J, Rapp M, Wang Q, et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020; 584(7821): 450-456.
[Crossref] [Google Scholar] [Pubmed]
- WHO. Coronavirus (COVID-19) Dashboard. World Health Organization. 2022.
- Starr TN, Greaney AJ, Dingens AS, Bloom JD. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. Cell Rep Med. 2021; 2(4): 100255.
[Crossref] [Google Scholar] [Pubmed]
- Cherian S, Potdar V, Jadhav S, Yadav P, Gupta N, Das M, et al. SARS-CoV-2 spike mutations, L452R, T478K, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. Microorganisms. 2021; 9: 1542.
[Crossref] [Google Scholar] [Pubmed]
- Mannar D, Saville JW, Zhu X, Srivastava SS, Berezuk AM, Tuttle KS, et al. SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex. Science. 2022; 375(6582): 760-764.
[Crossref] [Google Scholar] [Pubmed]
- Park M, Won J, Choi BY, Lee CJ. Optimization of primer sets and detection protocols for SARS-CoV-2 of coronavirus disease 2019 (COVID-19) using PCR and real-time PCR. Exp Mol Med. 2020; 52(6): 963-977.
[Crossref] [Google Scholar] [Pubmed]
- Kim JJ, Park HM, Kyoung AY, Park IK, Lim SK, Park BC. Inclusion of double helix structural oligonucleotide (STexS) results in an enhance of SNP specificity in PCR. Sci Rep. 2021; 11(1): 1-7.
[Crossref] [Google Scholar] [Pubmed]
- van Blargan LA, Errico JM, Halfmann PJ, Zost SJ, Crowe JE, Purcell LA, et al. An infectious SARS-CoV-2 B. 1.1. 529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat Med. 2022; 28(3): 490-495.
[Crossref] [Google Scholar] [Pubmed]
Author Info
Jae Jong Kim1, Hyoung-Min Park2, A Young Kyoung1, Si-Kyu Lim1*, Sun Ho Cha1, J Eugene Lee2 and Byoung Chul Park3*2Department of Biometrology, Korea Research Institute of Standards and Science, Daejeon, Republic of Korea
3Department of Disease Target Structure Research, Korea Research Institute of Bioscience and Biotechnology, Daejeon, Republic of Korea
Citation: Kim JJ: Compact Method of SARS-CoV-2 PCR Detection Enhances Overall Process of Diagnosing Delta and Omicron Variants
Received: 29-Aug-2022 Accepted: 23-Sep-2022 Published: 30-Sep-2022, DOI: 10.31858/0975-8453.13.9.630-634
Copyright: This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
ARTICLE TOOLS
- Dental Development between Assisted Reproductive Therapy (Art) and Natural Conceived Children: A Comparative Pilot Study Norzaiti Mohd Kenali, Naimah Hasanah Mohd Fathil, Norbasyirah Bohari, Ahmad Faisal Ismail, Roszaman Ramli SRP. 2020; 11(1): 01-06 » doi: 10.5530/srp.2020.1.01
- Psychometric properties of the World Health Organization Quality of life instrument, short form: Validity in the Vietnamese healthcare context Trung Quang Vo*, Bao Tran Thuy Tran, Ngan Thuy Nguyen, Tram ThiHuyen Nguyen, Thuy Phan Chung Tran SRP. 2020; 11(1): 14-22 » doi: 10.5530/srp.2019.1.3
- A Review of Pharmacoeconomics: the key to “Healthcare for All” Hasamnis AA, Patil SS, Shaik Imam, Narendiran K SRP. 2019; 10(1): s40-s42 » doi: 10.5530/srp.2019.1s.21
- Deuterium Depleted Water as an Adjuvant in Treatment of Cancer Anton Syroeshkin, Olga Levitskaya, Elena Uspenskaya, Tatiana Pleteneva, Daria Romaykina, Daria Ermakova SRP. 2019; 10(1): 112-117 » doi: 10.5530/srp.2019.1.19
- Dental Development between Assisted Reproductive Therapy (Art) and Natural Conceived Children: A Comparative Pilot Study Norzaiti Mohd Kenali, Naimah Hasanah Mohd Fathil, Norbasyirah Bohari, Ahmad Faisal Ismail, Roszaman Ramli SRP. 2020; 11(1): 01-06 » doi: 10.5530/srp.2020.1.01
- Manilkara zapota (L.) Royen Fruit Peel: A Phytochemical and Pharmacological Review Karle Pravin P, Dhawale Shashikant C SRP. 2019; 10(1): 11-14 » doi: 0.5530/srp.2019.1.2
- Pharmacognostic and Phytopharmacological Overview on Bombax ceiba Pankaj Haribhau Chaudhary, Mukund Ganeshrao Tawar SRP. 2019; 10(1): 20-25 » doi: 10.5530/srp.2019.1.4
- A Review of Pharmacoeconomics: the key to “Healthcare for All” Hasamnis AA, Patil SS, Shaik Imam, Narendiran K SRP. 2019; 10(1): s40-s42 » doi: 10.5530/srp.2019.1s.21
- A Prospective Review on Phyto-Pharmacological Aspects of Andrographis paniculata Govindraj Akilandeswari, Arumugam Vijaya Anand, Palanisamy Sampathkumar, Puthamohan Vinayaga Moorthi, Basavaraju Preethi SRP. 2019; 10(1): 15-19 » doi: 10.5530/srp.2019.1.3