Research Article - (2022) Volume 13, Issue 5
Abstract
The widespread of Acinetobacter baumanniiis concerning. The horizontal gene's transfer of this bacteria is crucial in acquiring unique traits like antibiotic resistance, which has been related to a significant increase in infection and fatality rates in patients. The New Delhi carbapenemase enzyme, which hydrolyzes all-lactam antibiotics (except aztreonam), including the broad-spectrum carbapenem antibiotic, is one of the most therapeutically significant carbapenemes. NDM (New Delhi metallo-beta lactamase) is a carbapenem enzyme that efficiently hydrolyzes lactams; as a result, NDM-producing bacteria only have a few treatment alternatives. VITEK-identified multidrug-resistant (MDR) A. baumannii isolates from Iraqi pneumonia patients were included in the investigation. Using 0.7 percent agarose gel electrophoresis, each isolate was checked for the presence of plasmid(s). Using the polymerase chain reaction technique, positive isolates were submitted to genetic identification of plasmid containing blaNDM1 and blaNMD2. The presence of the plasmid was discovered in all the isolates tested. NDM1-R1 enzyme was found to be positive in 61.5 percent of isolates, NDM1-R2 enzyme in 61.5 percent of isolates, and the NDM-2 enzyme in 61.5 percent of isolates. In addition, 38.4 percent of the participants tested negative for both enzymes. The importance of plasmid-mediated horizontal gene transfer in the acquisition of multiple drug resistance in MDR A. baumannii is highlighted by these findings. It is the first time to detect plasmid-mediated blaNDM1 and blaNDM2 genes in MDR A. baumannii in Iraq. The purpose of this study was to determine the incidence of NDM variants, metallo-β-lactamases, among A. baumannii isolates from diverse clinical samples in Iraq
Keywords
Acinetobacter baumannii, Plasmid, NDM-1, NDM-2
Introduction
Acinetobacter baumanniiis a Gram-negative, non-fermenting, aerobic, and nonmotile coccobacillus (Siroy A, et al., 2005). Currently, this bacterium is a typical occurrence in intensive care units and quickly became resistant to most antibiotics. Numerous outbreaks caused by this organism have been reported globally, (Huang YC, et al., 2002; Chan PC, et al., 2007; Hammerum AM, et al., 2015; Perez F, et al., 2007) including India, Egypt, and China (Chen Y, et al., 2011; Kaase M, et al., 2011; Karthikeyan K, et al., 2010).
Acinetobacter can cause Infections of the urinary tract, wounds, and burns (Fishbain J and Peleg AY, 2010). 3%-5% of nosocomial pneumonia cases are caused by it and are most common among patients in intensive care units (Dijkshoorn L, et al., 1993).
Due to A. baumannii’s proclivity to develop genes for resistance (Corbella X, et al., 2000), these infections are difficult to treat which yielded a multidrug-resistant (MDR) strain.1 This resistance developed as a result of the overuse of antibiotics (Peleg AY, et l., 2008). It is suggested that carbapenems can be used in treating patients and saving their lives (Gao L, et al., 2017). Carbapenems are the chosen medication to treat this pathogen (Costa LMD, et al., 2013).
The New Delhi metallo-β-lactamase 1 (blaNDM-1), also known as the carbapenem resistance gene, (Boulanger A, et al., 2012) was reported from a 59-year-old Swedish male in India. The man was hospitalized first in Punjab and then in New Delhi (Johnson AP and Woodford N, 2013). In January 2008, he was repatriated to Sweden, where an isolate of Klebsiella pneumoniae, which showed resistance to multiple antibiotics such as carbapenems, was obtained from his urine culture on the day after admission (Yong D, et al., 2009).
Carbapenem resistance was linked to bacteria's ability to propagate via mobile genetic elements, according to the phenotypic examination of isolates (Miriagou V, et al., 2010). The ISAba125 element, which is found upstream of the NDM-1 gene, may enhance the rapid dispersion of the NDM-1 gene; it is one of several factors that can influence NDM transmission (Poirel L, et al., 2012).
Carbapenem resistance is caused by a variety of causes. First, it is through the formation of hydrolyzing carbapenem-β-lactamases by A. baumannii (Hornsey M, et al., 2011). β-lactamases are enzymes produced by some of these bacteria that can make them resistant to various classes of β-lactam antibiotics, the main treatment for these infections. In the mid-1980s, a new group of these enzymes was detected, the extended-spectrum β-lactamases (ESBLs), which confer resistance to expanded-spectrum cephalosporins but not to carbapenems. Carbapenems are primarily used for treating infections due to ESBL-producing Enterobacteriaceae. These particular groups of β-lactamases are classified as class B (metallo-β-lactamases (MBLs)-like) and class D (oxacillinases), including OXA-23-like, OXA-24/40-like, and OXA- 58 (Ambler RP, et al., 1991). Second, it is through alterations in porin permeability in the outer membrane of the microorganism, in addition to efflux pumps and modifications in penicillin-binding protein affinity (Abbott I, et al., 2013). The blaNDM-1gene encodes an NDM-1 enzyme that is resistant to all-lactam antibiotics, including carbapenems, which have been discovered in Enterobacteriaceae. (Klebsiella pneumoniae and Escherichia coli) (Yong D, et al., 2009; Kumarasamy KK, et al., 2010) as well as in A. baumannii, where the blaNDM-1and blaNDM-2 genes were recently discovered (Espinal P, et al., 2013). The rapid evolution of blaNDM-1 has been linked to a mobile plasmid that may be passed from one bacteria to the next, from one person to the next, and even from one country to the next (Fallah F, et al., 2011).
The first NDM-2-producing A. baumannii isolate was discovered in the urine of an Egyptian woman brought to Tawam Hospital (Al Ain, UAE) in May 2009, after she had been hospitalized for cancer treatment in Cairo and Beirut several times the previous year. The second group of isolates was discovered in July 2009 during a thorough surveillance investigation conducted on patients admitted to two wards of the Tel Aviv-based TA-Sourasky-MA Rehabilitation Hospital (the Sourasky Medical Center) (Es-pinal P, et al., 2013).
Kaase M, et al. (Kaase M, et al., 2011) recently described an NDM-2 variation (proline to alanine substitution at position 28). This allele was originally discovered in an MDR A. baumannii strain isolated from a German patient who had previously been hospitalized in Egypt (Kaase M, et al., 2011), followed by another isolate from Israel.
These new findings inspired us to be the first to explore and investigate the transferable plasmid, which is thought to play a role in the dissemination of A. baumannii since many Iraqi patients have traveled to India and other nations for medical treatment in recent years, which may have aided in the acquisition of this gene.
Materials and Methods
Bacteria collection and identification
A. baumannii sample was collected from 39 sputum and bronchioalveolar lavage samples taken from patients suffering from pneumonia, admitted to Al-Yarmuk hospital from September 2019 until February 2020. Bacteria identification was achieved using the VITEK system. All confirmed specimens were cultured on Mueller-Hinton agar plates to test their susceptibility using the Kirby-Bauer disk diffusion method according to the described guidelines, which were suggested by the Clinical and Laboratory Standards Institute (CLSI).
The used antibiotics were ampicillin (MIC ≥ 32 mg/l), aztreonam and ceftriaxone (MIC ≥ 64 mg/l), gentamicin (MIC ≥ 16 mg/l), ciprofloxacin (MIC ≥ 4 mg/l), imipenem (MIC ≥ 16 mg/l), levofloxacin (MIC ≥ 8 mg/l), tigecycline (MIC ≥ 1 mg/l), and colistin (MIC ≥ 1 mg/l).
Identified bacteria were then stored at -80°C in nutrient broth with 20% glycerol. Isolates were cultured in Luria-Bertani broth for plasmid extraction.
Plasmid extraction and molecular detection of NDM-1 and NDM-2 genes
Extraction of plasmid from the isolates was done according to the manufacturer’s instructions (Wizard plus SV MInipreps DNA purification system, Promega, USA) with modification so that the elution buffer was minimized in the last step of plasmid DNA extraction to 35 µl. For plasmid detection, 5 µl of plasmid extract was mixed with 2 µl of loading dye (Promega, USA) and resolved on 0.7% gel electrophoresis, which was run at 70 V for 2 h. Then, the plasmid molecular size was specified according to a ladder, ranging 10,200-500 bp. (Bioneer, Republic of Korea).
For genetic detection of NDM-1 and NDM-2 genes, the Polymerase Chain Reaction (PCR) technique (PCR Express, Hybaid, USA) was applied. The primers used are listed in Table 1. Master Mix (ambGood, Canada) was applied according to the manufacturer’s instructions, and the total volume was 50 µl. Thermocycler conditions applied for both genes are listed in Table 2. PCR products were later resolved by 1.5% gel electrophoresis at 70 V for 1.5 h using a ladder ranging from 2000 to 100 bp (Bioneer, Republic of Korea).
| Gene | Primer sequence | Product size (bp) |
|---|---|---|
| NDM1-R1 | F:5-GGGCAGTCGCTTCCAACGGT-3 | 476 |
| R:5-GTAGTGCTCAGTGTCGGCAT-3 | ||
| NDM1-R2 | F:5-GGGCAGTCGCTTCCAACGGT-3 | 391 |
| R:5-GTGCCGTAGCTCCCAACGGT-3 | ||
| NDM2 | F:5-GTCGCAAAGCCCAGCTTCGCA-3 | 945 |
| R:5-GCCTCGCATTTGCGGGGTTTTTA-3 | ||
Table 1: List of primers used to amplify the genes encoding carbapenemases
| Gene | Initial denaturation | Denaturation | Annealing | Extension | Final extension | Number of cycles |
|---|---|---|---|---|---|---|
| NDM-1 | 95°C/10 min | 94°C/30 s | 60°C/30 s | 72°C/1 min | 72°C/7 min | 30 cycles |
| NDM-2 | 95°C/10 min | 94°C/30 s | 62°C/30 s | 72°C/1 min | 72°C/7 min | 30 cycles |
Table 2: PCR thermocycling conditions
Results
Confirmed positive bacteria were cultured on Mueller-Hinton agar plates to test their susceptibility using the Kirby-Bauer disk diffusion method; the rate of the resistance was different from one antibiotic to another. For ampicillin, the rate of resistance was 92%; aztreonam, 90%; cefotaxime, 87.2%; gentamicin, 72.5%; imipenem, 46%; levofloxacin, 45.1%; tigecycline, 41.4; and colistin, 27.2%.
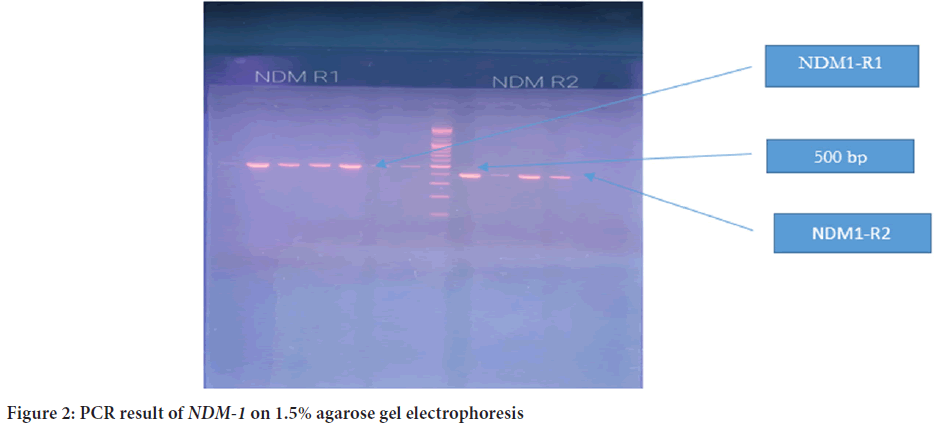
The plasmid was detected in all 39 isolates, and the molecular size was more than 10,200 bp (Figure 1). PCR was later conducted to detect the presence of blaNDM1 and blaNDM2 genes in the plasmid of 39 isolates. The PCR product sizes for both NDM1-R1 and NDM1-R2 were 476 bp and 391 bp, respectively (Figure 2). For NDM-2, the PCR product size was 946 bp (Figure 3). The results revealed that 24 out of 39 (61.5%) isolates demonstrated positivity for NDM1-R1 and NDM1-R2, while 15 out of 39 (38.4%) isolates showed negative results for these genes. For NDM2-R, 24 out of 39 (61.5%) isolates appeared to have this gene, whereas 15 out of 39 (38.4%) isolates showed negative results (Table 3). R1 and R2 (reverse primers): Referring to the used primer, which is a short sequence of single-stranded DNA that marks both ends of the target sequence; the reverse primer attaches to the stop codon of the complementary strand of DNA.
| Gene | No. of positive samples | Positive percentage | No. of negative samples | Negative percentage |
|---|---|---|---|---|
| NDM1-R1 | 24/39 | 61.50% | 15/39 | 38.40% |
| NDM1-R2 | 24/39 | 61.50% | 15/39 | 38.40% |
| NDM-2 | 24/39 | 61.50% | 15/39 | 38.40% |
Table 3: Number of positive and negative isolates harboring NDM-1 and NDM-2 genes
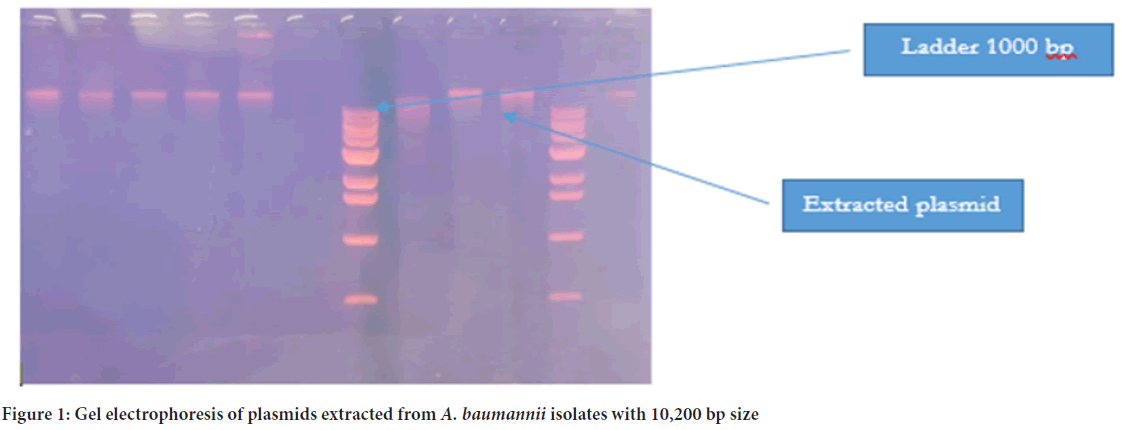
Figure 1: Gel electrophoresis of plasmids extracted from A. baumannii isolates with 10,200 bp size
Figure 2: PCR result of NDM-1 on 1.5% agarose gel electrophoresis

Figure 3: PCR result of NDM-2 on 1.5% agarose gel electrophoresis
Discussion
A. baumannii, a prominent nosocomial pathogen, is a serious public health concern, especially in intensive care units. Lactams, aminoglycosides, carbapenems, and fluoroquinolones are among the antimicrobials to which some strains of this bacterium are resistant to (Abbo A, et al., 2005). The growing multidrug resistance, which includes colistin and carbapenem, causes concern about this pathogen.
Even though these plasmids play a significant role and crucial part in A. baumannii resistance towards the antibiotic, due to their ability to transmit virulence and antimicrobial genes and despite their significant role in the pathogenicity of the bacteria, there has been no research into the role of plasmids in the transmission of NDM-1 and NDM-2 genes in Iraq.
Due to the quick disruption of the gene between Enterobacterales and Acinetobacter spp., NDM carbapenems have been identified in most parts of the world (Antunes LCS, et al., 2014). Only a few studies on NDM-1-producing A. baumannii have been published in Africa, and those that have been published are largely from northern or southern African countries, as well as Benin. Another study was done in Iraq by Sarah and Suhad in 2018 on Pseudomonas aeruginosa to detect these genes (Ismail SJ and Mahmoud SS, 2018). This is the first study on the dissemination of blaNDM-1 and blaNDM-2 genes in Al-Yarmuk hospitals among A. baumannii isolates (Ismail SJ and Mahmoud SS, 2018).
The results of antibiotic susceptibility exhibited different patterns of resistance to different types of antibiotics. Results showed resistance to at the very least, three types of antibiotics; As a result, the isolates were classified as MDR.
The isolates demonstrated the highest resistance to ampicillin (92%); these results are a little lower than another study conducted by Al-Harmoosh and Jarallah, (Al-Harmoosh RA and Jarallah EM, 2015) where they found that their resistance was 100% due to the wide range use of ampicillin in Yarmuk hospital. The present study also showed their high resistance to aztreonam (90%), and this result is close to another survey done by Alubaidi, et al. (Alubaidi GT, et al., 2021) where their result was 90.8. Also, the result was 80% in Al-Hilla hospital in a study by Al-Harmoosh and Jarallah, (Al-Harmoosh RA and Jarallah EM, 2015) and there was a moderate resistance to cefotaxime (87.2), which is in accordance with the result of Alubaidi, et al. (Alubaidi GT, et al., 2021) who noted that the resistance was 87.3, while the result of Al-Harmoosh and Jarallah (Al-Harmoosh RA and Jarallah EM, 2015) was (60%). The development of porin-deficient mutants may be a cause of resistance to cephalosporin antibiotics (Manchanda V and Singh NP, 2013). Furthermore, an increasing number of bacterial strains exhibit various forms of β-lactamases, including inducible and/or plasmid-mediated AmpC enzymes, which may raise the risk of cephalosporin resistance (Al-Harmoosh RA and Jarallah EM, 2015).
The resistance to gentamicin was 72.5, which is lower than the result of Alsehlawi and his colleagues, who found the resistance to be 83.3% (Alsehlawi ZS, et al., 2014). Other studies that were done in Thailand by Leepethacharat and Oberdorfer (Leepethacharat K and Oberdorfer P, 2007) showed a result of 60%; their result was in parallel with that of a study conducted in Turkey conducted by Özdemir and his colleagues (Özdemir H, et al., 2011). In A. baumannii, resistance to gentamicin is frequently caused by the production of modifying enzymes such as aminoglycoside-modifying enzymes (AMEs), acetylases, phosphorlyases, and adenylases, all of which can reduce antibiotic efficacy. Changes in bacterial membrane permeability and ribosomal protein alteration are two further resistance mechanisms (Barros JCS, et al., 1999). In the current study, the impedance against ciprofloxacin (quinolone antibiotic) was 68.7, while in a study by Al-Harmoosh and Jarallah, (Al-Harmoosh RA and Jarallah EM, 2015) it was 40%. In China, Zhou and his colleagues discovered that clinical isolates were resistant to ciprofloxacin in a significant percentage of cases (>95%) (Zhou H, et al., 2007). Alsehlawi and his colleagues discovered 91.6 percent resistance to ciprofloxacin in another investigation in Najaf (Alsehlawi ZS, et al., 2014). Quinolone resistance is caused by mutations in type II topoisomerases that are chromosomally encoded, as well as overexpression of efflux pumps or point-related genes (Drlica K and Zhao X, 1997; Tran JH, et al., 2005). The plasmid qnr genes are playing an increasingly important role in the spread of fluoroquinolone resistance (Firoozeh F, et al., 2014).
A low resistance rate to imipenem was recorded (46.5%), which is convergent with another study done by Talib, et al. (Talib ST, et al., 2018) while for levofloxacin, it was 45.1%, which is close to the result of Alubaidi, et al. (Alubaidi GT, et al., 2021) but contrast to the result obtained in a study done in Brazil, (Tognim MCB, et al., 1999) where it was 20%, as well as the outcome of Patwardhan's and his colleagues' efforts in India (Patwardhan RB, et al., 2008). This could be related to the A. baumannii-carrying multi-resistant plasmid (Patwardhan RB, et al., 2008). The lowest resistance was to colistin, with a rate of 27.2%, which corresponds with another study in which 97% of the bacteria were sensitive to colistin (Tunyapanit W, et al., 2014).
The current study's high levels of antibiotic resistance could be due to both innate and acquired mechanisms. Resistance is widespread, posing a significant clinical risk (Mathur P, et al., 2002). Antibiotics are also subjected to selection pressure in the hospital setting, resulting in diverse resistances to these medications. According to El-Astal (El-Astal Z, 2005), the increasing resistance rate of A. baumannii to routinely used antimicrobial medications could be due to inadequate and erroneous antimicrobial agent administration, as well as a lack of suitable infection management methods (El-Astal Z, 2005).
Recently, we have witnessed an increase in resistance to imipenem and other antibiotics in our hospitals; this resistance is because of this organism’s ability to horizontally acquire resistance deterrents. Resistance manifests itself in a variety of ways, including the production of enzymes such as β-lactamases, which hydrolyze the β-lactam group and carbapenem. AMEs are enzymes that modify aminoglycosides.
In the present study, plasmid with a size of more than 10,200 bp was detected in A. baumannii. These results coincide with another study in Baghdad conducted by Alubaidi, et al. (Alubaidi GT, et al., 2021) in Al-Khadmiya Hospital.
In 2012, in China, isolates from a clinical Acinetobacter lwoffii strain were studied (Hu H, et al., 2012). According to the researchers, the plasmid had a complete Tn125 transposon and had a strong horizontal transferability, where they found one large plasmid with a size of 78,125 bp and three smaller ones with sizes of 4797, 1634, and 7865 bp, respectively (Galata V, et al., 2015).
Since then, several blaNDM-1positive plasmids have been discovered in Acinetobacter spp in China. In A. baumannii, blaNDM-1was discovered on plasmids ranging in size from 30 to 50 kb in a Chinese investigation. NDM-1 was discovered predominantly on plasmids in Acinetobacter spp in china. China, while it was found on the bacterial chromosomes in Europe and North Africa (Hu H, et al., 2012; Zhou Z, et al., 2012) Jones et al. discovered NDM-1 on numerous bands ranging in size from 45 to 300 kb in an Indian investigation (Jones LS, et al., 2014).
Other studies reported that A. baumannii possesses plasmids ranging from as small as 2 kb to more than 100 kb in size (Gallagher LA, et al., 2015; Lean SS and Yeo CC, 2017). Due to the inclusion of several antibiotic resistance genes and their self-transmissible merit, A. baumannii big plasmids are frequently the focus of the investigation (Hamidian M and Hall RM, 2018). Small plasmids, particularly those harboring antibiotic resistance genes, were emphasized in other studies (Gallagher LA, et al., 2015; Lean SS and Yeo CC, 2017).
A. baumannii that produces NDM-1 was first discovered in India (Karthikeyan K, et al., 2010) and then in China (Chen Y, et al., 2011). Non-baumannii species that produce NDM-1 Acinetobacter genomospecies have been documented from nosocomial settings and environmental sources in China, including Acinetobacter lwoffii, Acinetobacter junii, Acinetobacter pittii, Acinetobacter haemolyticus, and Acinetobacter haemolyticus.
Experiments and molecular research confirmed that the NDM-1 gene is localized on transferable plasmids of 180 and 140 kb belonging to the pNDM-BJ01-like family, in contrast to NDM-1-producing A. baumannii, the blaNDM-2's reservoir (Hu H, et al., 2012). Plasmids in K. pneumoniae and E. coli isolates are frequently conjugative, which aids in the transmission of genes across strains of different genera (Johnson AP and Woodford N, 2013; Espinal P, et al., 2011).
Following the sequencing of the NDM-1-bearing plasmid, a fragment of ISAba125 was discovered just upstream of the NDM-1 gene (Poirel L, et al., 2011). According to several researches, the NDM gene is located between two copies of the ISAba125 element, generating the Tn125 composite transposon. These transposons can mobilize a wide range of resistance genes, aiding in the spread of antimicrobial resistance (Mahillon J and Chandler M, 1998). These transpositions are one of the most common causes of bacterial DNA rearrangements, which can result in gene expression alterations (Sinha MH, 2004). Transposable elements (ISAba125 element) give the 35 sequences of the hybrid promoter in A. baumannii, which is responsible for NDM gene production (Poirel L, et al., 2011). In A.baumannii, the genes ISAba1,ISAba2,ISAba3,ISAba4, and IS18 are frequently related with carbapenemase gene expression (Villalón P, et al., 2013).
Only one NDM-1 variant (NDM-2) was detected in A. baumannii isolates, and it was described in 2011 by Kaase, et al. It differed by a single amino acid (Kaase M, et al., 2011). Following that, Espinal, et al. (Espinal P, et al., 2011) sequenced the NDM gene, finding a twofold nucleotide alteration from cytosine to guanine at position 82 and alanine to guanine at position 468 from the start codon. Only the first modification, which was earlier published and called NDM-2, resulted in an amino acid substitution from P (proline) to A (alanine) at position 28, and the other was a quiet mutation. This variation has also been found in a German patient (who had previously traveled to Egypt) and in a patient from the United Arab Emirates (Ghazawi A, et al., 2012). In 2010, NDM-1 and NDM-2 were found in A.baumannii, and in 2011, NDM-5 was found in E.coli(Hornsey M, etal., 2011). In 2018, Sarah and Suhad detected NDM-1and NDM-2among Pseudomonasaeruginosa(Ismail SJ and Mahmoud SS, 2018).
In the current study, NDM-1 and NDM-2 genes were harbored by a plasmid with a size ranging from approximately 10,200 bp, which can be conjugated and transmitted to E. coli Although these investigations were conducted in vitro, we could predict that the NDM genes were spread from Acinetobacter species to Enterobacteriaceae because of this plasmid's interspecies transfer (Bonnin RA, et al., 2014).
The first case of an MDR A. baumannii strain generating NDM-1 and NDM-2 in Iraq is described here. The possibility of variations developing will increase if this bacterium containing the NDM-1 and NDM-2 genes continues to spread. This is a crucial factor to consider when developing genetic tools to target carbapenem resistance genes, especially because many Iraqi patients seek treatment in India, which could make getting these genes easier.
We were inspired by these new results to be the first to research and explore the transferable plasmid, which is assumed to play a role in the spread of A. baumannii. In recent years, many Iraqi patients have sought medical treatment in India and other countries, which may have contributed to the acquisition of this gene.
Conclusion
As in the present study, isolates with the NDM-positive A. baumannii gene that produces New Delhi metallo-β-lactamase exhibit resistance to most antimicrobials recommended by CLSI (Clinical and Laboratory Standards Institute, 2012) and appear to be MDR, especially against ampicillin, where the resistance rate was 92%, and this may create a serious problem in choosing therapy. This New Delhi metallo-β-lactamase, which is produced by the NDM-1 gene, can be a target for a new therapy.
References
- Siroy A, Molle V, Guillier LC, Vallenet D, Caron PM, Cozzone AJ, et al. Channel formation by CarO, the carbapenem resistance-associated outer membrane protein of Acinetobacter baumannii. Antimicrob Agents Chemother. 2005; 49(12): 4883-4876.
[Crossref] [Google scholar] [Pubmed]
- Huang YC, Su LH, Wu TL, Hsieh WS, Chang TM, Lin TY. Outbreak of Acinetobacter baumanni bacteraemia in neonatal intensive care unit: Clinical implications and genotyping analysis. Pediatr Infect Dis J. 2002; 21: 1105-1109.
- Chan PC, Huang LM, Lin HC, Chang LY, Chen ML, Liu CY, et al. Control of an outbreak of pandrug-resistant Acinetobacter baumannii colonization and infection in neonatal intensive care unit. Infect Control Hosp Epidemiol. 2007; 28: 423-429.
[Crossref] [Google scholar] [Pubmed]
- Hammerum AM, Hansen F, Skov MN, Stegger M, Andersen PS, Holm A, et al. Investigation of a possible outbreak of carbapenem-resistant Acinetobacter baumanniiin Odense, Denmark using PFGE, MLST and whole-genome-based SNPs. J Antimicrob Chemother. 2015; 70: 1965-1968.
[Crossref] [Google scholar] [Pubmed]
- Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2007; 51: 3471-3484.
[Crossref] [Google scholar] [Pubmed]
- Chen Y, Zhou Z, Jiang Y, Yu Y. Emergence of NDM 1-producing Acinetobacter baumannii in China. J Antimicrob Chemother. 2011; 66(6): 1255-1259.
[Crossref] [Google scholar] [Pubmed]
- Kaase M, Nordmann P, Wichelhaus TA, Gatermann SG, Bonnin RA, Poirel L. NDM-2 carbapenemase in Acinetobacter baumanniifrom Egypt. J Antimicrob Chemother. 2011; 66, 1260-1262.
[Crossref] [Google scholar] [Pubmed]
- Karthikeyan K, Thirunarayan MA, Krishnan P. Coexistence of blaOXA-23 with blaNDM 1 and armA in clinical isolates of Acinetobacter baumanniifrom India. J Antimicrob Chemother. 2010; 65(10): 2253-2254.
[Crossref] [Google scholar] [Pubmed]
- Fishbain J, Peleg AY. Treatment of acinetobacter infections. Clin Infect Dis. 2010; 51(1): 79-84.
[Crossref] [Google scholar] [Pubmed]
- Dijkshoorn L, Aucken HM, Smidt GP, Kaufmann ME, Ursing J, Pitt TL. Correlation of typing methods for acinetobacter isolates from hospital outbreaks. J Clin Microbiol. 1993; 31: 702-705.
[Crossref] [Google scholar] [Pubmed]
- Corbella X, Montero A, Pujol M, Dominguez MA, Ayats J, Argerich MJ, et al. Emergence and rapid spread of carbapenem resistance during a large and sustained hospital outbreak of multi-resistant Acinetobacter baumannii. J Clin Microbiol. 2000; 38: 4086-4095.
[Crossref] [Google scholar] [Pubmed]
- Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: Emergence of a successful pathogen. Clin Microbiol Rev. 2008; 21(3): 538-582.
[Crossref] [Google scholar] [Pubmed]
- Gao L, Lyu Y, Li Y. Trends in drug resistance of Acinetobacter baumanniiover a 10-year period: Nationwide data from the China surveillance of antimicrobial resistance program. Chin Med J. 2017; 130(6): 659-664.
[Crossref] [Google scholar] [Pubmed]
- Costa LMD, Coelho JM, Souza HA, Castro ME, Stier CJ, Bragagnolo KL, et al. Outbreak of carbapenem-resistant Acinetobacter baumanniiproducing the OXA-23 enzyme in Curitiba, Brazil. J Clin Microbiol. 2003; 41: 3403-3406.
[Crossref] [Google scholar] [Pubmed]
- Boulanger A, Naas T, Fortineau N, Figueiredo S, Nordmann P. NDM 1 producing Acinetobacter baumanniifrom Algeria. Antimicrob Agents Chemother. 2012; 56(4): 2214-2215.
[Crossref] [Google scholar] [Pubmed]
- Johnson AP, Woodford N. Global spread of antibiotic resistance: The example of New Delhi metallo-β-lactamase (NDM)-mediated carbapenem resistance. J Med Microbiol. 2013; 62(4): 499-513.
[Crossref] [Google scholar] [Pubmed]
- Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, et al. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009; 53(12): 5046-5054.
[Crossref] [Google scholar] [Pubmed]
- Miriagou V, Cornaglia G, Galani I, Giske CG, Gniadkowski M, Lada EM, et al. Acquired carbapenemases in gram-negative bacterial pathogens: Detection and surveillance issues. Clin Microbiol Infect. 2010; 16(2): 112-122.
[Crossref] [Google scholar] [Pubmed]
- Poirel L, Bonnin RA, Boulanger A, Schrenzel J, Kaase M, Nordmann P. Tn125 related acquisition of blaNDM like genes in Acinetobacter baumannii. Antimicrob Agents Chemother. 2012; 56(2): 1087-1089.
[Crossref] [Google scholar] [Pubmed]
- Hornsey M, Phee L, Wareham DW. A novel variant NDM-5 of the New Delhi metallo-β-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob Agents Chemother. 2011; 5(11): 5952-5954.
[Crossref] [Google scholar] [Pubmed]
- Ambler RP, Coulson AF, Frére JM, Ghuysen JM, Joris B, Forsman M, et al. A standard numbering scheme for the class A β-lactamases. Biochem J. 1991; 276: 269-270.
[Crossref] [Google scholar] [Pubmed]
- Abbott I, Cerqueira GM, Bhuiyan S, Peleg AY. Carbapenem resistance in Acinetobacter baumannii: Laboratory challenges, mechanistic insights and therapeutic strategies. Expert Rev Antiinfect Ther. 2013; 11: 395-409.
[Crossref] [Google scholar] [Pubmed]
- Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infect. Dis. 2010; 10: 597-602.
[Crossref] [Google scholar] [Pubmed]
- Espinal P, Poirel L, Carmeli Y, Kaase M, Pal T, Nordmann P, et al. Spread of NDM-2-producing Acinetobacter baumannii in the Middle East. J Antimicrob Chemother. 2013; 68(8): 1928-1930.
[Crossref] [Google scholar] [Pubmed]
- Fallah F, Taherpour A, Vola MH, Hashemi A. Global spread of New Delhi metallo-beta-lactamase-1(NDM-1). Iran J Clin Infect Dis. 2011; 6(4): 171-176.
- Abbo A, Venezia NS, Muntz HO, Krichali T, Igra SY, Carmeli Y. Multidrug resistant Acinetobacter baumannii. Emerg Infect Dis. 2005; 11(1): 22-29.
[Crossref] [Google scholar] [Pubmed]
- Antunes LCS, Visca P, Towner KJ. Acinetobacter baumannii: Evolution of a global pathogen. Pathog Dis. 2014; 71: 292-301.
[Crossref] [Google scholar] [Pubmed]
- Ismail SJ, Mahmoud SS. First detection of New Delhi metallo-β-lactamases variants (NDM-1, NDM-2) among Pseudomonas aeruginosa isolated from Iraqi hospitals. Iran J Microbiol. 2018; 10(2): 98-103.
[Google scholar] [Pubmed]
- Al-Harmoosh RA, Jarallah EM. First detection of the blaNDM-1 and blaNDM-2 genes in a clinical isolates of Acinetobacter baumanniiin Hillah hospitals-Iraq. Int J Adv Res. 2015; 1407-1416. [Crossref]
[Google scholar] [Pubmed]
- Alubaidi GT, Jamal QW, Abdulrahman TR, Jasim IA, Ahmed IW. Detection of plasmid-mediated OqxA and OqxB genes in clinical isolates of Acinetobacter baumannii taken from Iraqi pneumonic patients. Proceeding of the IIER International Conference, Melbourne, Australia. 2021.
- Manchanda V, Singh NP. Occurrence and detection of AmpC β-lactamases among Gram-negative clinical isolates using a modified three-dimensional test at Guru Tegh Bahadur Hospital, Delhi, India. J Antimicrob Chemother. 2003; 51(2): 415-418.
[Crossref] [Google scholar] [Pubmed]
- Alsehlawi ZS, Alshara JM, Hadi ZJ, Almohana AM. First report of the blaoxa-23 gene in a clinical isolates of Acinetobacter baumanniiin Najaf hospitals-Iraq. Int J Recent Sci Res. 2014; 5(8): 1407-1411.
- Leepethacharat K, Oberdorfer P. Acinetobacter baumannii infection and colonization among pediatric patients at Chiang Mai University Hospital. J Infect Dis Antimicrob Agents. 2007; 24(2): 63-73.
- Özdemir H, Kendirli T, Ergün H, Çiftçi E, Tapısız A, Güriz H, et al. Nosocomial infections due to Acinetobacter baumanniiin a pediatric intensive care unit in Turkey. Turk J Pediatr. 2011; 53: 255-260. [Crossref]
[Google scholar] [Pubmed]
- Barros JCS, Bozza M, Filho GFJ, Bello AB, Lopes UG, Pereira JAA. Evidences of gentamicin resistance amplification in Klebsiella pneumoniae isolated from faeces of hospitalized newborns. Mem Inst Oswaldo Cruz Rio de Janeiro. 1999; 94(6): 795-802.
- Zhou H, Pi BR, Yang Q, Yu YS, Chen YG, Li LJ, et al. Dissemination of imipenem-resistant Acinetobacter baumanniistrains carrying the ISAba1 blaOXA-23 genes in a Chinese hospital. J Med Microbiol. 2007; 56: 1076-1080.
[Crossref] [Google scholar] [Pubmed]
- Drlica K, Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev. 1997; 61: 377-392.
[Crossref] [Google scholar] [Pubmed]
- Tran JH, Jacoby GA, Hooper DC. Interaction of the plasmid encoded quinolone resistance protein qnr with Escherichia coli DNA gyrase. Antimicrob Agents Chemother. 2005; 49(1): 118-125.
[Crossref] [Google scholar] [Pubmed]
- Firoozeh F, Zibaei M, Soleimani-Asl Y. Detection of plasmid-mediated qnr genes among the quinolone-resistant Escherichia coli isolates in Iran. J Infect Dev Ctries. 2014; 8(7): 818-822.
[Crossref] [Google scholar] [Pubmed]
- Talib ST, Abdulrahman TR, Ali SH. Detection of some biofilm genes related with MDR in A. baumannii isolated from clinical isolates. Iraqi JMS. 2018; 16(4): 430-438.
- Tognim MCB, Gaziri LCJ, Vidotto MC, Perugini MR. Association of plasmid typing to biotyping and antibiotyping in the characterization of outbreaks by Acinetobacter baumannii. Braz Arch Biol Technol. 1999; 42(1): 1-8.
- Patwardhan RB, Dhakephalkar PK, Niphadkar KB, Chopade BA. A study on nosocomial pathogens in ICU with special reference to multi-resistant Acinetobacter baumannii harbouring multiple plasmids. Indian J Med Res. 2008; 128(2): 178-187.
[Google scholar] [Pubmed]
- Tunyapanit W, Pruekprasert P, Laoprasopwattana K, Chelae S. Antimicrobial susceptibility of Acinetobacter baumanniiisolated from hospital patients. Science Asia. 2014; 40: 28-34.
- Mathur P, Tatman A, Das B, Dhawan B. Prevalence of extended beta lactamase producing gram negative bacteria in a tertiary care hospital. Indian J Med Res. 2002; 115: 153-157.
[Pubmed]
- El-Astal Z. Increasing ciprofloxacin resistance among prevalent urinary tract bacterial isolates in Gaza Strip, Palestine. J Biomed Biotechnol. 2005; (3): 238-241.
[Crossref] [Google scholar] [Pubmed]
- Hu H, Hu Y, Pan Y, Liang H, Wang H, Wang X, et al. Novel plasmid and its variant harboring both a blaNDM-1 gene and type IV secretion system in clinical isolates of Acinetobacter lwoffii. Antimicrob Agents Chemother. 2012; 56: 1698-1702.
[Crossref] [Google scholar] [Pubmed]
- Galata V, Fehlmann T, Backes C, Keller A. PLSDB: A resource of complete bacterial plasmids. Nucleic Acids Res. 2015; 47: D195-D202.
[Crossref] [Google scholar] [Pubmed]
- Zhou Z, Guan R, Yang Y, Chen L, Fu J, Deng Q, et al. Identification of New Delhi metallo-beta-lactamase gene (NDM-1) from a clinical isolate of Acinetobacter junii in China. Can J Microbiol. 2012; 58: 112-115.
[Crossref] [Google scholar] [Pubmed]
- Jones LS, Toleman MA, Weeks JL, Howe RA, Walsh TR, Kumarasamy KK. Plasmid carriage of blaNDM-1 in clinical Acinetobacter baumanniiisolates from India. Antimicrob Agents Chemother. 2014; 58(7): 4211-4213.
[Crossref] [Google scholar] [Pubmed]
- Gallagher LA, Ramage E, Weiss EJ, Radey M, Hayden HS, Held KG, et al. Resources for genetic and genomic analysis of emerging pathogen Acinetobacter baumannii. J Bacteriol. 2015; 197: 2027-2035.
[Crossref] [Google scholar] [Pubmed]
- Lean SS, Yeo CC. Small, enigmatic plasmids of the nosocomial pathogen, Acinetobacter baumannii: Good, bad, who knows? Front Microbiol. 2017; 8: 1-8.
- Hamidian M, Hall RM. Genetic structure of four plasmids found in Acinetobacter baumannii isolate D36 belonging to lineage 2 of global clone 1. PLoS One. 2018; 13: 1-15.
[Crossref] [Google scholar] [Pubmed]
- Espinal P, Fugazza G, Lopez Y, Kasma M, Lerman Y, Kumar MS, et al. Dissemination of an NDM-2-producing Acinetobacter baumanniiclone in an Israeli rehabilitation center. Antimicrob Agents Chemother. 2011; 55, 5396-5398.
[Crossref] [Google scholar] [Pubmed]
- Poirel L, Bonnin RA, Nordmann P. Analysis of the resistome of a multidrug-resistant NDM-1-producing Escherichia coli strain by high-throughput genome sequencing. Antimicrob Agents Chemother. 2011; 55: 4224-4229.
[Crossref] [Google scholar] [Pubmed]
- Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998; 62, 725-774.
[Crossref] [Google scholar] [Pubmed]
- Sinha MH. Genes VIII Lewin. 2004: 467-480.
- Villalón P, Valdezate S, Pascual MMJ, Carrasco G, Vindel A, Nieto SJA. Epidemiology of the Acinetobacter-derived cephalosporinase: Carbapenem-hydrolysing oxacillinase and metallo-β-lactamase genes, and of common insertion sequences, in epidemic clones of Acinetobacter baumannii from Spain. J Antimicrob Chemother. 2013; 68: 550-553.
[Crossref] [Google scholar] [Pubmed]
- Ghazawi A, Sonnevend A, Bonnin RA, Poirel L, Nordmann P, Hashmey R, et al. NDM-2 carbapenemase-producing Acinetobacter baumannii in the United Arab Emirates. Clin Microbiol Infect. 2012; 18(2): e34-e36.
[Crossref] [Google scholar] [Pubmed]
- Bonnin RA, Poirel L, Nordman P. New Delhi metallo-β-lactamase-producing Acinetobacter baumannii: A novel paradigm for spreading antibiotic resistance genes. Future Microbiol. 2014; 9(1): 33-41.
[Crossref] [Google scholar] [Pubmed]
Author Info
Dalyia Akram Hamoodi*Citation: Hamoodi DA: Detection of Plasmid-Mediated blaNDM1 and blaNDM2 Genes in Clinical Isolates of Acinetobacter baumannii from Iraqi Patients
Received: 01-Apr-2022 Accepted: 29-Apr-2022 Published: 06-May-2022, DOI: 10.31858/0975-8453.13.5.342-348
Copyright: This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
ARTICLE TOOLS
- Dental Development between Assisted Reproductive Therapy (Art) and Natural Conceived Children: A Comparative Pilot Study Norzaiti Mohd Kenali, Naimah Hasanah Mohd Fathil, Norbasyirah Bohari, Ahmad Faisal Ismail, Roszaman Ramli SRP. 2020; 11(1): 01-06 » doi: 10.5530/srp.2020.1.01
- Psychometric properties of the World Health Organization Quality of life instrument, short form: Validity in the Vietnamese healthcare context Trung Quang Vo*, Bao Tran Thuy Tran, Ngan Thuy Nguyen, Tram ThiHuyen Nguyen, Thuy Phan Chung Tran SRP. 2020; 11(1): 14-22 » doi: 10.5530/srp.2019.1.3
- A Review of Pharmacoeconomics: the key to “Healthcare for All” Hasamnis AA, Patil SS, Shaik Imam, Narendiran K SRP. 2019; 10(1): s40-s42 » doi: 10.5530/srp.2019.1s.21
- Deuterium Depleted Water as an Adjuvant in Treatment of Cancer Anton Syroeshkin, Olga Levitskaya, Elena Uspenskaya, Tatiana Pleteneva, Daria Romaykina, Daria Ermakova SRP. 2019; 10(1): 112-117 » doi: 10.5530/srp.2019.1.19
- Dental Development between Assisted Reproductive Therapy (Art) and Natural Conceived Children: A Comparative Pilot Study Norzaiti Mohd Kenali, Naimah Hasanah Mohd Fathil, Norbasyirah Bohari, Ahmad Faisal Ismail, Roszaman Ramli SRP. 2020; 11(1): 01-06 » doi: 10.5530/srp.2020.1.01
- Manilkara zapota (L.) Royen Fruit Peel: A Phytochemical and Pharmacological Review Karle Pravin P, Dhawale Shashikant C SRP. 2019; 10(1): 11-14 » doi: 0.5530/srp.2019.1.2
- Pharmacognostic and Phytopharmacological Overview on Bombax ceiba Pankaj Haribhau Chaudhary, Mukund Ganeshrao Tawar SRP. 2019; 10(1): 20-25 » doi: 10.5530/srp.2019.1.4
- A Review of Pharmacoeconomics: the key to “Healthcare for All” Hasamnis AA, Patil SS, Shaik Imam, Narendiran K SRP. 2019; 10(1): s40-s42 » doi: 10.5530/srp.2019.1s.21
- A Prospective Review on Phyto-Pharmacological Aspects of Andrographis paniculata Govindraj Akilandeswari, Arumugam Vijaya Anand, Palanisamy Sampathkumar, Puthamohan Vinayaga Moorthi, Basavaraju Preethi SRP. 2019; 10(1): 15-19 » doi: 10.5530/srp.2019.1.3