Review Article - (2024) Volume 15, Issue 9
Mechanism of Antibiotic Resistance by Protein Moonlighting
Ikeanyibe Nneoma Collette*, Sabinus Oscar O. Eze, Tobechukwu Christian Ezike and Kingsley Ozioma OmejeAbstract
Antibiotic resistance has become a major global health crisis, posing significant challenges in the treatment of bacterial infections. Most antibiotic resistance has emerged as a result of mutation or through the transfer of genetic material between microorganisms. Some studies on bacterial resistance have shown that there is a huge diversity of resistance mechanisms, with their distribution and interaction being mostly complex and unknown. However, various biochemical and physiological mechanisms are responsible for the development of antibiotic resistance. However, emerging evidence suggests that protein moonlighting, a phenomenon where a single protein can perform multiple functions in the cell beyond its originally described role, plays an important role in antibiotic resistance. Some proteins involved in the normal functioning of bacterial cells may have the ability to interact with antibiotics in ways that allow the bacteria to resist their effects. For example, Pyruvate Dehydrogenase (PDH), a key enzyme in energy metabolism, can bind to and detoxify fluoroquinolone antibiotics like ciprofloxacin. In Pseudomonas aeruginosa (P. aeruginosa), PDH sequesters the antibiotic within its active site, rendering it inactive. Additionally, some proteins involved in the transport of nutrients into the bacterial cell may also be able to transport antibiotics out of the cell, preventing them from accumulating to toxic for the bacteria. For example, ToIC an outer membrane protein found in Escherichia coli (E. coli) act as a channel for various molecules, including antibiotics. However, specific mutations in ToIC can alter its conformation, reducing the passage of certain antibiotics like cephalosporin’s and there by offering resistance. Other proteins may modify or degrade antibiotics within the cell, rendering them ineffective. For example the protein enolase, which is involved in glycolysis, has been shown to possess β-lactamase activity, allowing it to inactivate β lactam antibiotics. Overall, protein moonlighting is a complex phenomenon that can contribute to antibiotic resistance in a various ways. Understanding how these proteins function in bacterial cells and how they interact with antibiotics is an important area of research that could ultimately lead to the development of novel strategies for combating antibiotic-resistant bacteria.
Keywords
Protein moonlighting, Antibiotics, Antibiotic resistance, Glycolysis, Mutation, Pyruvate dehydrogenase
Introduction
From the Plague of Athens during the Peloponnesian War (430 BC), which killed approximately one third of the population (Huremovic D, 2019 ), to the Corona Virus Disease pandemic (COVID-19), which resulted in 14.9 million deaths (WHO, 2022 ), the massive impact of infectious illnesses has led to pandemics and outbreaks resulting in countless fatalities across the global population (Spellberg B, et al., 2008), Antimicrobial agents have created to reduce infection-related mortality. For example, santonin, cathelicidins and aminoglycosides are effective antibacterial substances obtained from plants, animals and microbes (Chin KW, et al., 2023). Antibiotics are class of antimicrobial agents used to treat bacterial infections. They work by targeting specific cellular processes or structures in bacteria, which either kill the bacteria directly bactericidal or prevent them from growing and proliferating bacteriostatic (Brown ED and Wright GD, 2016). When antibiotics are consumed, enzymes in the body degrade them and convert into active substance that inhibit the microbial growth, there by curing the infection. However, the excretion of antibiotic residues or un-metabolized antibiotics in urine or feaces, along with their entry into the environment through sewage systems, cause high levels of antibiotics in the environment (Martinez JL, 2008). Excessive antibiotics in the ecosystem force bacteria to undergo a selection process that causes mutation of pathogens and the development of immunity to existing antibiotics (Kollef MH, et al., 2017 ). These antibiotic resistant bacteria known as superbugs, develop antibiotic resistance genes for carrying out resistance mechanisms against antibiotics. For example, Methicillin-Resistant Staphylococcus aureus (MRSA) has mutated the methicillin-resistant structural gene (MecA gene or Staphylococcal Cassette Chromosome mec (SCCmec) gene) against methicillin inhibition. In 2019, MRSA caused over 100,000 infections, in making it one of the greatest threats to human health.
Antibiotic resistance occurs when bacteria evolve to become resistant to the effects of antibiotics, making them difficult or impossible to treat. This can happen when antibiotics are used too frequently or inappropriately, allowing bacteria to develop mechanisms that resist their effects (Levy SB and Marshall B, 2004). Antibiotic resistance can make common bacterial infections, such as pneumonia, tuberculosis and urinary tract infections, much more difficult to treat leading to increased morbidity, mortality and healthcare costs.
Addressing the problem of antibiotic resistance requires a multifaceted approach that includes the appropriate use of antibiotics, the development of antibiotics and measures to prevent the spread of resistant bacteria and the development of alternative therapies such as phage therapy and immunotherapy (Ventola CL, 2015).
Literature Review
Traditional mechanism of antibiotic resistance
Bacteria have the ability to develop resistance to antibiotics through various mechanisms. The mechanism of antibiotic resistance varies depending on the type of antibiotic and the bacteria involved. However, there are several ways that bacteria can become resistant to antibiotics including
Mutation: Bacteria can mutate in a way that make them resistant to antibiotics. For example, a mutation might cause the bacteria to produce an enzyme that inactivates the antibiotic, or alter the structure of the target site to which the antibiotic would normally bind, preventing it from antibiotic from working effectively (Davies J and Davies D, 2010). For example, mutations in the target protein of antibiotics like fluoroquinolones, can reduce the binding affinity of the drug, rendering it ineffective (Hooper DC, 1999).
Acquisition of resistance genes: One of the most common mechanisms of antibiotic resistance is through the acquisition of resistance genes. Bacteria can acquire these genes from other bacteria, through a process called horizontal gene transfer, where genetic material such as plasmids, intergrons or transposons containing antibiotic resistance genes is exchanged (Bush K, et al., 2011). For instance, the widespread dissemination of Extended Spectrum β-Lactamase (ESBL) genes is mainly attributed to plasmid-mediated gene transfer (Pitout JD, 2012).
Efflux pumps: Some bacteria can become resistant to antibiotics by using efflux pumps, which are proteins that pump antibiotics out of the bacterial cell before they can exert their effects (Blair JM, et al., 2015).
Biofilm formation: Another mechanism of antibiotic resistance is through the formation of biofilms. A biofilm is a protective matrix that can make bacteria more resistant to antibiotics, as this matrix can prevent the antibiotics from reaching the bacteria (Flemming HC, et al., 2016).
Enzymatic inactivation: Some bacteria produce enzymes that can modify or degrade antibiotics, rendering them inactive. For example, β-lactamase enzymes hydrolyze β-lactam such as penicillins and cephalosporins, leading to their inactivation (Drawz SM and Bonomo RA, 2010) (Figure 1).
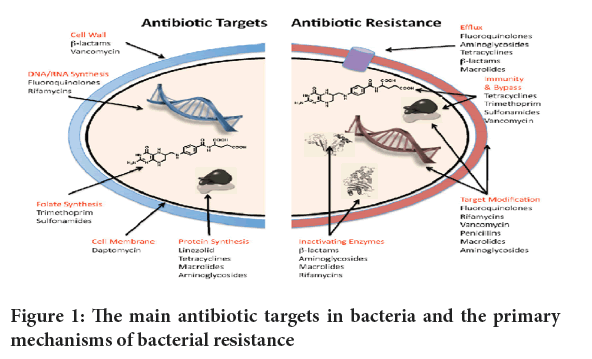
Figure 1: The main antibiotic targets in bacteria and the primary mechanisms of bacterial resistance.
Protein moonlighting
Protein moonlighting is a phenomenon in which a single protein can perform multiple unrelated functions in the cell, in addition to its primary or canonical function. These alternative functions are often distinct from the proteins primary function and may not be immediately apparent from its sequence or structure. This concept challenges the traditional view of one gene, one protein, one function proposed by Beadle GW and Tatum EL, 1941.
Moonlighting proteins are a subclass of multifunctional proteins that have numerous physiologically significant biochemical or biophysical functions through a single polypeptide chain (Jeffery CJ, 2003). Nearly 1000 proteins have been identified as having a second function, with enzymes accounting for about two-thirds of them (Jeffery CJ, 2014; Franco SL, et al., 2021). Numerous types of proteins, such as receptors, enzymes, transcription factors, adhesions and scaffolds are examples of moonlighting proteins and various functional combinations. Moonlighting proteins are expressed throughout the evolutionary tree and function in many different biochemical pathways. Some can perform both functions simultaneously, while for others the proteins function changes in response to environment conditions. Protein moonlighting has been observed in various organisms and plays a significant role in cellular processes. This phenomenon provides a unique adaptive advantage to organisms, allowing them to maximize the utility of a limited number of genes (Jeffery CJ, 2015). For example, certain metabolic enzymes have been found to have additional functions in processes such as signal transduction, cytoskeletal organization and transcriptional regulation (Copley SD, 2012). Furthermore, protein moonlighting enables the development of intricate regulatory networks and the integration of diverse cellular processes (Wistow G, 2011).
The diverse examples of moonlighting proteins already identified, along with the potential benefits they may provide to organism, suggest that many more moonlighting proteins are likely to be found (Jeffery CJ, 2018). Continuing studies of the structures and functions of these proteins will aid in predicting the functions of proteins identified through genome sequencing projects, in interpreting results from proteomics experiments, in understanding how different biochemical pathways interact in systems biology, in annotating protein sequence and structure databases, in studies of protein evolution and in the design of proteins with novel functions (Jeffery CJ, 2018).
Moonlighting proteins in biological systems
Metabolic enzymes: Some moonlighting proteins are enzymes involved in metabolic pathways that also have non-enzymatic functions (Krantz M and Klipp E, 2020). A notable example is Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) is a key enzyme in glycolysis but has been found to have additional roles in DNA repair, transcriptional regulation and apoptosis (Dadgar A, et al., 2019; Drlica K and Zhao X, 1997). Lactate dehydrogenase, an enzyme of the glycolytic pathway which, can also function as a structural component of the lens of the eye as shown in Figure 2.
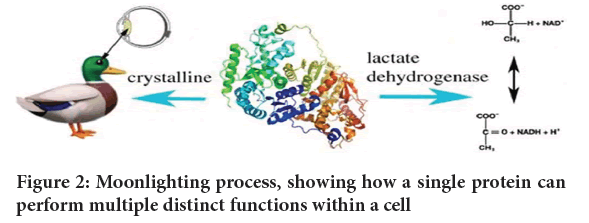
Figure 2: Moonlighting process, showing how a single protein can perform multiple distinct functions within a cell.
Chaperones: Chaperones are proteins that help other proteins in folding correctly and have been found to have moonlighting functions. For instance, Heat Shock Proteins (HSPs), a type of chaperone protein particularly important in helping cells to cope with stress, such as heat shock or exposure to toxins and can also regulate gene expression, interact with signaling proteins and modulate immune responses (Vos MJ, et al., 2017).
Structural proteins: Some proteins involved in maintaining cellular structure and integrity have been shown to possess moonlighting functions. For example actin is a major component of the cytoskeleton, can also participate in intracellular transport, gene regulation and signaling pathways (Kim JH and Kim JJ, 2017).
Virulence factors: In pathogenic microorganisms, moonlighting proteins can play important roles in virulence. For instance, some bacterial adhesions and toxins have been found to have additional functions in host immune modulation or cellular invasion (Henderson B and Martin A, 2011; Huberts DH and van der Klei IJ, 2010). Additionally, certain viral proteins can moonlight as host proteins, which can allow the virus to evade the host’s immune system (Henderson B and Martin A, 2010).
Mechanisms of protein moonlighting
Mechanisms of protein moonlighting involve various processes and molecular interactions that enable a single protein to perform multiple distinct functions including:
Gene sharing: A single gene can encode multiple proteins, each with a different function. For example, the gene for lactate dehydrogenase can encode a protein that functions as an enzyme in the glycolytic pathway, as well as a protein that functions as a structural component.
Post-translational modification: A protein can be modified after it is translated, through processes such as acetylation, phosphorylation or glycosylation. These modifications can change the proteins structure and function, allowing it to perform multiple tasks. For example, the protein subunit alpha) from the antibiotic isoniazid, contributing to antibiotic resistance.
Impact of protein moonlighting in antibiotic efficacy
Protein moonlighting has a significant impact on antibiotic efficacy (de la Cruz F et al., 2017) For example, a protein that serves as an antibiotic target may function as a drug resistance factor. This can make it difficult to develop antibiotics that are effective against bacteria that have evolved resistance to existing drugs.
A specific example of how protein moonlighting can impact antibiotic efficacy is the case of the protein Secretion ATPase (SecA). SecA is essential for protein secretion in bacteria and also acts as an antibiotic target. Some antibiotics, such as bacitracin, work by binding to SecA and preventing its function. However, bacteria can evolve resistance to bacitracin by mutating the SecA protein, rendering it an unavailable target for the antibiotic (Liu X, et al., 2019).
Another example of how protein moonlighting can impact antibiotic efficacy is the case of the protein GroEL (Heat shock protein 60 or (Hsp60)). GroEL is essential for protein folding in bacteria and also functions as a moonlighting protein that can act as a drug resistance factor. Some antibiotics, such as ciprofloxacin, work by binding to GroEL and preventing it from functioning. However, bacteria can evolve resistance to ciprofloxacin by mutating the GroEL protein, rendering it a less effective target for the antibiotic (de la Cruz et al., 2017).
Conclusion
Protein moonlighting, a phenomenon where a single protein performs multiple distinct functions within a cell, has emerged as a novel mechanism in antibiotic resistance. Understanding how moonlighting proteins contribute to resistance mechanisms can guide the design of novel therapeutics that specifically target these proteins or their interactions with antibiotics. For example, researchers may explore the development of compounds that disrupt the moonlighting function of a protein involved in resistance while leaving its primary function intact. The impact of protein moonlighting on antibiotic efficacy is a complex issue; however, it is clear that protein moonlighting can play a significant role in the development of antibiotic resistance. As we continue to develop antibiotics, it is important to consider the potential for protein moonlighting to impact their efficacy. Moonlighting proteins, which have multiple functions, are attractive targets for antibiotic development, as they could be used to disrupt multiple pathways in the bacteria, making it more difficult for the bacteria to develop resistance.
References
- Huremovic D. Brief history of pandemics throughout history. Psychiatry of pandemics: A mental health response to infection outbreak. 2019: 7-35.
- World Health Organization (WHO). 14.9 million excess deaths associated with the COVID-19 pandemic in 2020 and 2021. 2022.
- Spellberg B, Guidos R, Gilbert D, Bradley J, Boucher HW, Scheld WM, et al. The epidemic of antibiotic-resistant infections: A call to action for the medical community from The Infectious Diseases Society Of America. Clin Infect Dis. 2008; 46(2): 155-164.
[Crossref] [Google Scholar] [PubMed]
- Chin KW, Tiong HL, Luang V, Ma NL. An overview of antibiotic and antibiotic resistance. Environ Adv. 2023; 11: 100331.
- Brown ED, Wright GD. Antibacterial drug discovery in the resistance era. Nature. 2016; 529(7586): 336-343.
[Crossref] [Google Scholar] [PubMed]
- Martínez JL. Antibiotics and antibiotic resistance genes in natural environments. Science. 2008; 321(5887): 365-367.
[Crossref] [Google Scholar] [PubMed]
- Kollef MH, Bassetti M, Francois B, Burnham J, Dimopoulos G, Garnacho MJ, et al. The intensive care medicine research agenda on multidrug-resistant bacteria, antibiotics and stewardship. Intensive Care Med. 2017; 43: 1187-1197.
[Crossref] [Google scholar] [Pubmed]
- Levy SB, Marshall B. Antibacterial resistance worldwide: Causes, challenges and responses. Nat Med. 2004; 10(12): 122-129.
[Crossref] [Google scholar] [Pubmed]
- Ventola CL. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm Therap. 2015; 40(4): 277.
- Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010; 74(3): 417-433.
[Crossref] [Google scholar] [Pubmed]
- Hooper DC. Mechanisms of fluoroquinolone resistance. Drug Resist Updat. 1999; 2(1): 38-55.
[Crossref] [Google scholar] [Pubmed]
- Bush K, Courvalin P, Dantas G, Davies J, Eisenstein B, Huovinen P, et al. Tackling antibiotic resistance. Nat Rev Microbiol. 2011; 9(12): 894-896.
[Crossref] [Google Scholar] [PubMed]
- Pitout JD. Extraintestinal pathogenic Escherichia coli: An update on antimicrobial resistance, laboratory diagnosis and treatment. Expert Rev Anti Infect Ther. 2012; 10(10): 1165-1176.
[Crossref] [Google Scholar] [PubMed]
- Blair JM, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJ. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. 2015; 13(1): 42-51.
[Crossref] [Google Scholar] [PubMed]
- Flemming HC, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. Biofilms: An emergent form of bacterial life. Nat Rev Microbiol. 2016; 14(9): 563-575.
[Crossref] [Google Scholar] [PubMed]
- Drawz SM, Bonomo RA. Three decades of β-lactamase inhibitors. Clin Microbiol Rev. 2010; 23(1): 160-201.
[Crossref] [Google Scholar] [PubMed]
- Beadle GW, Tatum EL. Genetic control of biochemical reactions in Neurospora. Proc Natl Acad Sci. 1941; 27(11): 499-506.
[Crossref] [Google Scholar] [PubMed]
- Jeffery CJ. Moonlighting proteins: Old proteins learning new tricks. Trends Genet. 2003; 19(8): 415-417.
[Crossref] [Google Scholar] [PubMed]
- Jeffery CJ. An introduction to protein moonlighting. Biochem Soc Trans. 2014; 42(6): 1679-1683.
[Crossref] [Google Scholar] [PubMed]
- Franco SL, Sanchez RD, Najar GA, Hernandez S, Amela I, Pinol J, et al. Pathogen moonlighting proteins: From ancestral key metabolic enzymes to virulence factors. Microorganisms. 2021; 9(6): 1300.
[Crossref] [Google Scholar] [PubMed]
- Jeffery CJ. Why study moonlighting proteins? Front Genet. 2015; 6: 211.
- Copley SD. Moonlighting enzymes: Proteins with multiple functions in cells. Handbook of Molecular Microbial Ecology I: Metagenomics and Complementary Approaches. 2012;147-156.
- Wistow G. Moonlighting proteins: The insulation of biological functions. Trends Cell Biol. 2011; 21(9): 522-531.
- Jeffery CJ. Protein moonlighting: What is it, and why is it important? Philos Trans R Soc Lond B Biol Sci. 2018; 373(1738): 20160523.
[Crossref] [Google Scholar] [PubMed]
- Krantz M, Klipp E. Moonlighting proteins an approach to systematize the concept. In Silico Biol. 2020; 13(3-4): 71-83.
[Crossref] [Google Scholar] [PubMed]
- Dadgar A, Asghari SM, Rezaei GN. Moonlighting function of glyceraldehyde-3-phosphate dehydrogenase: New mechanistic insights from multifunctional moonlighting proteins.Biochem Biophys Res Commun. 2019; 520(2): 412-416.
- Drlica K, Zhao X. DNA gyrase, topoisomerase IV and the 4-quinolones. Microbiol Mol Biol Rev. 1997; 61(3): 377-392.
[Crossref] [Google Scholar] [PubMed]
- Vos MJ, Hageman J, Carra S, Kampinga HH. Structural and functional diversities between members of the human HSPB, HSPH, HSPA and DNAJ chaperone families. Biochemistry. 2008; 47(27): 7001-7011.
- Kim JH, Kim JJ. Post-translational modifications of actin: A comprehensive review. Int J Biol Sci. 2017; 13(2): 121-139.
- Henderson B, Martin A. Bacterial moonlighting proteins and bacterial virulence. Curr Top Microbiol Immunol. 2011:155-213.
[Crossref] [Google Scholar] [PubMed]
- Huberts DH, van der Klei IJ. Moonlighting proteins: An intriguing mode of multitasking. Biochim Biophys Acta Mol Cell Res. 2010; 1803(4): 520-525.
[Crossref] [Google Scholar] [PubMed]
- de la Cruz F, Garcia MJ, Del campo FJ. Protein moonlighting: A threat to antibiotic efficacy. Nat Rev Microbiol. 2017; 15(1): 47-59.
- Liu X, Fan Y, Zhang C, Dai M, Wang X, Li W. Nuclear import of a secreted Candidatus Liberibacter asiaticusprotein is temperature dependent and contributes to pathogenicity in Nicotiana benthamiana. Front Microbiol. 2019; 10: 1684.
[Crossref] [Google Scholar] [PubMed]
Author Info
Ikeanyibe Nneoma Collette*, Sabinus Oscar O. Eze, Tobechukwu Christian Ezike and Kingsley Ozioma OmejeCitation: Collette IN: Mechanism of Antibiotic Resistance by Protein Moonlighting
Received: 03-Sep-2024 Accepted: 19-Sep-2024 Published: 26-Sep-2024, DOI: 10.31858/0975-8453.15.9.283-286
Copyright: This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
ARTICLE TOOLS
- Dental Development between Assisted Reproductive Therapy (Art) and Natural Conceived Children: A Comparative Pilot Study Norzaiti Mohd Kenali, Naimah Hasanah Mohd Fathil, Norbasyirah Bohari, Ahmad Faisal Ismail, Roszaman Ramli SRP. 2020; 11(1): 01-06 » doi: 10.5530/srp.2020.1.01
- Psychometric properties of the World Health Organization Quality of life instrument, short form: Validity in the Vietnamese healthcare context Trung Quang Vo*, Bao Tran Thuy Tran, Ngan Thuy Nguyen, Tram ThiHuyen Nguyen, Thuy Phan Chung Tran SRP. 2020; 11(1): 14-22 » doi: 10.5530/srp.2019.1.3
- A Review of Pharmacoeconomics: the key to “Healthcare for All” Hasamnis AA, Patil SS, Shaik Imam, Narendiran K SRP. 2019; 10(1): s40-s42 » doi: 10.5530/srp.2019.1s.21
- Deuterium Depleted Water as an Adjuvant in Treatment of Cancer Anton Syroeshkin, Olga Levitskaya, Elena Uspenskaya, Tatiana Pleteneva, Daria Romaykina, Daria Ermakova SRP. 2019; 10(1): 112-117 » doi: 10.5530/srp.2019.1.19
- Dental Development between Assisted Reproductive Therapy (Art) and Natural Conceived Children: A Comparative Pilot Study Norzaiti Mohd Kenali, Naimah Hasanah Mohd Fathil, Norbasyirah Bohari, Ahmad Faisal Ismail, Roszaman Ramli SRP. 2020; 11(1): 01-06 » doi: 10.5530/srp.2020.1.01
- Manilkara zapota (L.) Royen Fruit Peel: A Phytochemical and Pharmacological Review Karle Pravin P, Dhawale Shashikant C SRP. 2019; 10(1): 11-14 » doi: 0.5530/srp.2019.1.2
- Pharmacognostic and Phytopharmacological Overview on Bombax ceiba Pankaj Haribhau Chaudhary, Mukund Ganeshrao Tawar SRP. 2019; 10(1): 20-25 » doi: 10.5530/srp.2019.1.4
- A Review of Pharmacoeconomics: the key to “Healthcare for All” Hasamnis AA, Patil SS, Shaik Imam, Narendiran K SRP. 2019; 10(1): s40-s42 » doi: 10.5530/srp.2019.1s.21
- A Prospective Review on Phyto-Pharmacological Aspects of Andrographis paniculata Govindraj Akilandeswari, Arumugam Vijaya Anand, Palanisamy Sampathkumar, Puthamohan Vinayaga Moorthi, Basavaraju Preethi SRP. 2019; 10(1): 15-19 » doi: 10.5530/srp.2019.1.3