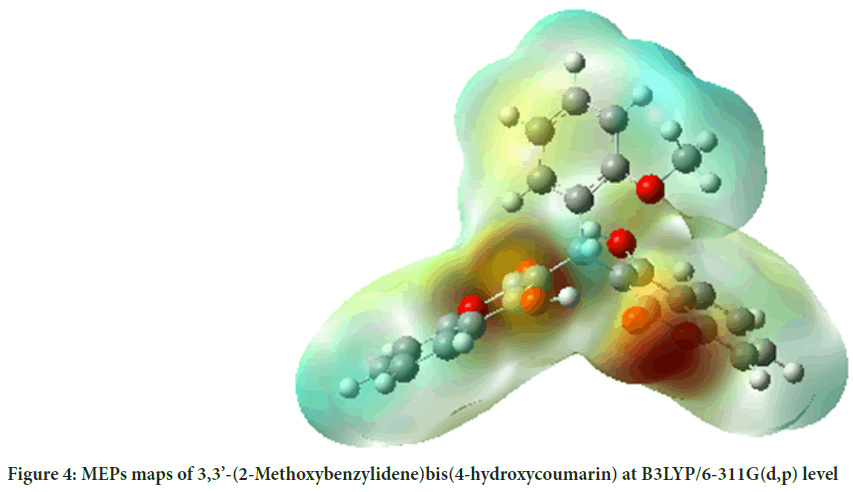Research Article - (2024) Volume 15, Issue 2
Abstract
In this study, the geometric structure determination of 3,3’-(2-Methoxybenzylidene)bis(4-hydroxycoumarin) molecule and its parameters (bond length and angles), Nuclear Magnetic Resonance (NMR) spectroscopes, electrical (HOMO, HOMO-1, LUMO and LUMO+1) properties, Mulliken, NBO and Atomic Polar Tensor (APT) charges and Molecular Electrostatic Potential (MEP) surfaces have been theoretically calculated utilizing the Gaussian 09 software. All theoretical calculations were calculated by using B3LYP (Becke’s 3-parameter hybrid functional using B exchange and LYP correlation) and HSEH1PBE (The exchange part of the screened Coulomb potential of Heyd, Scuseria, and Ernzerhof) levels of Density Functional Theory (DFT) method with 6-311++G(d,p) basis set. Moreover, DNA interactions have been viewed employing the molecular docking method of 3,3’-(2-Methoxybenzylidene) bis(4-hydroxycoumarin) molecule with the COVID-19 main protease (PDB 6LU7) responsible for the replication of corona virus.
Keywords
COVID-19, Molecular docking, DFT, NMR spectra
Introduction
The coronavirus emerged as a rapidly spreading epidemic in the city of Wuhan, China, in late 2019. An effective antiviral drug has not yet been developed against the SARS-CoV-2 virus, which was named as a global epidemic on March 11, 2020 by the World Health Organization. Rapid and detailed research is ongoing as there is an urgent need to search for effective antiviral agents to combat the COVID-19 virus. Today, many researches go on in drug design using Molecular Docking (MD) studies. Protein-ligand interaction studies by Molecular Docking (MD) play an important role in the knowledge of mechanisms in the discovery, design and development of drugs. Ligand selection in molecular docking is based on their antiviral activity.
The natural coumarins play an important role in plant biochemistry and physiology. They act as antioxidants, enzyme inhibitors and precursors of toxic substances. They are also involved in the actions of plant growth hormones and growth regulators, the control over the respiration and photosynthesis, as well as in the defense against various infections (Weinmann I, 1997). Although most of the existing natural coumarins have been isolated from higher plants, some of them have been discovered in microorganisms, e.g., aminocoumarin antibiotics: Novobiocin, coumermycin A1 and chlorobiocin (produced by the actinomycete Streptomyces niveus) (Marcu MG, 2000).
Synthetic coumarin derivatives have been obtained by chemical modification of the coumarin ring. As a substitution can conceptually occur at any of the six available sites of the basic molecule, these compounds are widely variable in structure and activity. The biological activities of coumarin derivatives, in particular their therapeutic application as anticoagulant and antibacterial agents (Borges F, 2005), has stimulated further interest for the synthesis of this class of compounds. A variety of synthesized coumarin derivatives have been experimentally shown to exert pharmacological activities including inhibition of platelet aggregation, cytochrome P450, and steroid 5-α-reductase. They have also been shown to exert efficient anti-proliferative, antifungal, anti-psoriasis, anti-inflammatory, as well as antiviral activities (Chiang CC, 2008; Cavar S, 2009; Jung JC, 2009; Symeonidis T, 2009; Kostova I, 2007). The interest in coumarins has recently increased significantly because it was found that they inhibit HIV (Human Immunodeficiency Virus), by affecting integrase and reverse transcriptase, which play a critical role in the replicative cycle of HIV (Nolan AK, 2009; Mahajan DH, 2009; Zhao H, et al., 1997). The present study is focused on the antiviral activity evaluation of the 3,3’-(2-Methoxybenzylidene)bis(4-hydroxycoumarin). In the light of this information, 3,3’-(2-Methoxybenzylidene)bis(4-hydroxycoumarin) molecule, one of the compounds containing thiophene with antiviral activity, may be among the drugs tested for the treatment of COVID-19 diseases. In order to better understand the molecular definition of the 3,3’-(2-Methoxybenzylidene) bis(4-hydroxycoumarin) molecule, first of all, the optimized structures of the molecules were determined using the Density Functional Theory (DFT) method in the Gaussian 09W program. Using optimized constructs in molecular docking calculations is more precise, making the program more reliable for use in construct-based drug design.
Materials and Methods
DFT calculations
The theoretical modeling was calculated for the 3,3’-(2-Methoxybenzylidene)bis(4-hydroxycoumarin) molecule by Gaussian 09 software (Frisch MJ, et al., 2009) in the ground state. The molecular structure of the optimized molecule was drawn by Gaussian View 5 program (Dennington R, 2009). Theoretical calculations of the 3,3’-(2-Methoxybenzylidene) bis(4-hydroxycoumarin) molecule performed with B3LYP (Becke AD, 1997; Lee C, 1988) and HSEh1PBE (Heyd J, 2004; Heyd J, 2004; Heyd J, 2005, Heyd J, 2003) with 6-311++G(d,p) (Frisch MJ, 1984). UV-vis was performed with TD-HSEh1PBE and TDB3LYP approach in ground-state.
Molecular docking calculations (ligand and target protein preparation)
Before starting the molecular docking calculations, the 3D molecular structure of COVID-19 (PDB 6LU7) were downloaded from the Protein Data Bank (PDB) of the Research Collaboratory for Structural Bioinformatics (RCSB) (Protein Data Bank, 2023). Thus, the ligand (3,3’-(2-Methoxybenzylidene)bis(4-hydroxycoumarin)) and target (PDB 6LU7) were determined for molecular docking calculations. Molecular Docking calculations were performed by using the AutoDock Tools (ADT) version 1.5.6 (Sanner MF, 1999) to find the ligand-protein docking interactions. The PyMOL software package (Delano WL, 2002) has analyzed the output of the AutoDock (version 4.0) program (Morris GM, 2009). In addition, discovery studio visualizer 3.5 software (BIOVIA discovery studio, 2024) was used to visualize the docked active sites in the protein and its H-bond interactions.
Results and Discussion
Geometric structure
The experimental (Završnik D, et al., 2011) structure of 3,3’-(2-Methoxybenzylidene)bis(4-hydroxycoumarin)molecule was shown in Figure 1.
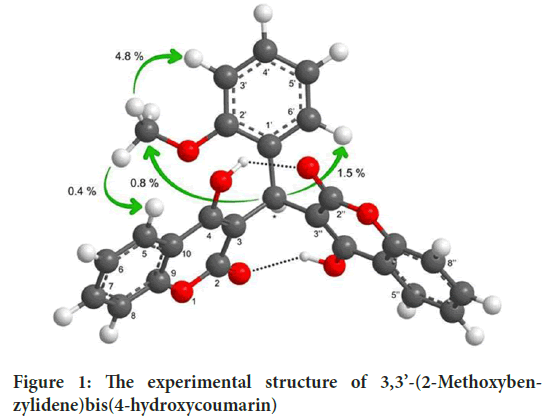
Figure 1: The experimental structure of 3,3’-(2-Methoxybenzylidene)bis(4-hydroxycoumarin)
Obtained geometric data listed in Table 1. In the study, Theoretical bond lengths (for C-C) were found in the range 1.36614-1.45154 Å and 1.35761-1.44606 (in Å) at B3LYP and HSEh1PBE, respectively. Experimental bond lengths (C-C) are seen in the range of 1.33508 and 1.45125 (in Å) (Završnik D, et al., 2011). Experimental O1-C2 bond lengths are 1.34557 Å, respectively (Završnik D, et al., 2011). Calculated bond lengths were observed as 1.37087 Å for the B3LYP and 1.36011 Å for the HSEh1PBE. THE experimental C9-O1-C2 bond angle is 121.07733˚ (Završnik D, et al., 2011), and this angle has been seen at 121.69770˚ for the B3LYP and 121.59628˚ for the HSEh1PBE.
| Parameters | Experimental | Theoretical | |
|---|---|---|---|
| Bond length (Å) | X-ray | DFT/B3LYP | DFT/HSEh1PBE |
| O1-C2 | 1.34557 | 1.37087 | 1.36011 |
| C2-C3 | 1.45125 | 1.44294 | 1.43642 |
| C3-C4 | 1.33508 | 1.37396 | 1.37114 |
| C4-C10 | 1.47082 | 1.45154 | 1.44606 |
| C10-C9 | 1.36542 | 1.39896 | 1.39526 |
| C9-O1 | 1.38919 | 1.36614 | 1.35761 |
| C9-C8 | 1.37677 | 1.39363 | 1.39014 |
| C8-C7 | 1.35041 | 1.38775 | 1.38430 |
| C7-C6 | 1.40660 | 1.40157 | 1.39788 |
| C6-C5 | 1.40437 | 1.38528 | 1.38178 |
| C5-C10 | 1.37807 | 1.40554 | 1.40090 |
| C1’-C2’ | 1.38504 | 1.41467 | 1.40987 |
| C2’-C3’ | 1.38100 | 1.39516 | 1.39157 |
| C3’-C4’ | 1.37617 | 1.39462 | 1.39115 |
| C4’-C5’ | 1.35959 | 1.38698 | 1.38384 |
| C5’-C6’ | 1.36939 | 1.39648 | 1.39272 |
| C6’-C1’ | 1.38859 | 1.39272 | 1.38867 |
| C2’’-C3’’ | 1.42824 | 1.44131 | 1.43580 |
| Bond angles ( ̊) | |||
| O1-C2-C3 | 119.31544 | 119.21389 | 119.38489 |
| C2-C3-C4 | 119.45233 | 119.52552 | 119.69162 |
| C3-C-C3’’ | 112.52285 | 112.11446 | 111.90222 |
| C-C3’’-C2’’ | 117.54075 | 119.76933 | 119.50576 |
| C3-C4-C10 | 120.30707 | 119.80790 | 119.40176 |
| C4-C10-C5 | 123.64224 | 123.24460 | 123.05636 |
| C4-C10-C9 | 117.46040 | 118.12013 | 118.13498 |
| C10-C9-O1 | 121.58466 | 121.18358 | 121.36507 |
| C9-O1-C2 | 121.07733 | 121.69770 | 121.59628 |
| C10-C9-C8 | 123.04578 | 121.55848 | 121.40624 |
| C9-C8-C7 | 118.44013 | 118.88060 | 118.89863 |
| C8-C7-C6 | 121.11161 | 120.57748 | 120.64294 |
| C7-C6-C5 | 118.82608 | 120.10227 | 120.03365 |
| C6-C5-C10 | 119.66419 | 120.24284 | 120.20647 |
| C5-C10-C9 | 118.89693 | 118.63427 | 118.80772 |
| C1’-C2’-C3’ | 121.34067 | 120.95104 | 120.86899 |
| C2’-C3’-C4’ | 119.88476 | 119.97973 | 119.94189 |
| C3’-C4’-C5’ | 119.30764 | 119.97013 | 120.04320 |
| C4’-C5’-C6’ | 121.13338 | 119.65885 | 119.59612 |
| C5’-C6’-C1’ | 120.97280 | 121.94429 | 121.87009 |
| C6’-C1’-C2’ | 117.33317 | 117.43165 | 117.60051 |
Table 1: Selected theoretical and experimental geometrical parameters
NMR spectroscopy
The combined use of NMR and computer simulation methods offers a powerful way to predict and interpret the structure of large biomolecules. In this study, The 13C and 1H NMR chemical shifts calculations have been carried out using the B3LYP and HSEH1PBE methods with 6-311++G(d,p) basis set for the optimized geometry. In Table 2, the experimental and the theoretical 1H and 13C isotropic chemical shifts (with respect to TMS, all values in ppm) for the 3,3’-(2-Methoxybenzylidene)bis (4-hydroxycoumarin)molecule has been given.
| Atom | Exp | B3LYP /6-311++G(d,p) | HSEh1PBE/6-311++G(d,p) |
|---|---|---|---|
| 1H | |||
| OCH3 | 3.57 | 3.33 | 3.40 |
| OCH3 | 3.57 | 3.40 | 3.49 |
| OCH3 | 3.57 | 3.78 | 3.86 |
| H* | 6.25 | 5.89 | 6.08 |
| H5' | 6.84 | 6.61 | 6.85 |
| H3' | 6.89 | 7.04 | 7.25 |
| H6 | 7.16 | 7.30 | 7.52 |
| H4' | 7.17 | 7.33 | 7.57 |
| H6 | 7.32 | 7.41 | 7.62 |
| H6" | 7.32 | 7.41 | 7.63 |
| H8 | 7.36 | 7.48 | 7.69 |
| H8" | 7.36 | 7.48 | 7.72 |
| H7 | 7.58 | 7.53 | 7.77 |
| H7" | 7.58 | 7.53 | 7.77 |
| H5 | 7.90 | 7.54 | 7.86 |
| H5" | 7.90 | 7.65 | 7.86 |
| 13C | |||
| C* | 32.93 | 38.12 | 31.45 |
| OCH3 | 55.51 | 54.19 | 48.50 |
| C3 | 104.80 | 111.44 | 104.13 |
| C3" | 104.80 | 111.44 | 104.13 |
| C3' | 110.94 | 114.60 | 107.76 |
| C8 | 115.91 | 120.58 | 115.65 |
| C8" | 115.91 | 120.58 | 115.65 |
| C10 | 117.54 | 122.58 | 116.26 |
| C10" | 117.54 | 123.29 | 116.92 |
| C5' | 119.85 | 123.91 | 118.65 |
| C5 | 123.60 | 127.85 | 122.69 |
| C5" | 123.60 | 127.85 | 122.92 |
| C4' | 127.29 | 128.14 | 127.96 |
| C6' | 128.22 | 129.43 | 129.54 |
| C1' | 128.29 | 129.73 | 130.96 |
| C7 | 131.63 | 131.33 | 131.90 |
| C7" | 131.63 | 132.87 | 131.90 |
Table 2: The calculated and experimental 13C and 1H isotropic NMR chemical shifts (with respect to TMS, all values in ppm
Electronic properties
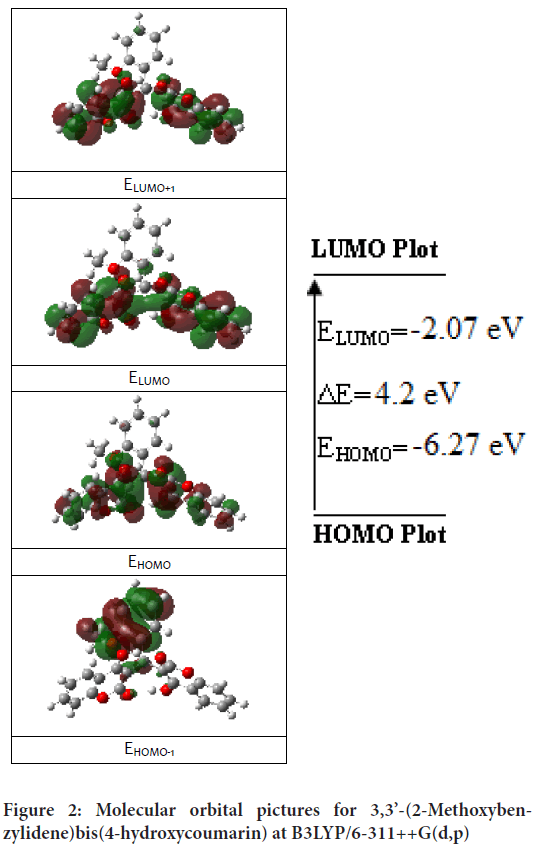
The transition of an electron from HOMO (Highest Occupied Molecular Orbit) to LUMO (Lowest Unoccupied Molecular Orbit) is defined as electronic absorption. Total energies of the frontier molecular orbital of the 3,3’-(2-Methoxybenzylidene)bis(4-hydroxycoumarin) molecule were investigated at the B3LYP method and shown in Figure 2. The energy gap is important in definition electrical transport properties of the molecule. Quantum chemical parameters (as chemical activity, HOMO-LUMO energies and their energy gap (ΔE), hardness (h), electronegativity (c), and electronic transition energies) were performed with B3LYP and HSEh1PBE methods and calculated values listed in Table 3.
| B3LYP /6-311++G(d,p) | HSEh1PBE /6-311++G(d,p) | |
|---|---|---|
| EHOMO(eV) | -6.27091 | -6.07336 |
| ELUMO(eV) | -2.06727 | -2.22727 |
| ΔE=ELUMO-EHOMO(eV) | 4.20364 | 3.84609 |
| I(eV) | 6.27091 | 6.07336 |
| A(eV) | 2.06727 | 2.22727 |
| c(eV) | 4.16909 | 4.150315 |
| h(eV) | 2.10182 | 1.923045 |
| S(eV-1) | 0.079733 | 0.082327 |
| ETOTAL(a.u) | -1528.61818533 | -1527.02554268 |
Table 3: FMOs, energies and calculated physico-chemical properties
Figure 2: Molecular orbital pictures for 3,3’-(2-Methoxybenzylidene)bis(4-hydroxycoumarin) at B3LYP/6-311++G(d,p)
The frontier orbital picture was depicted in Figure 2 with the 3D plots for the gas phase, and the positive and negative phases are represented in red and green color, respectively.
Charge analysis and Molecular Electrostatic Potential (MEP)
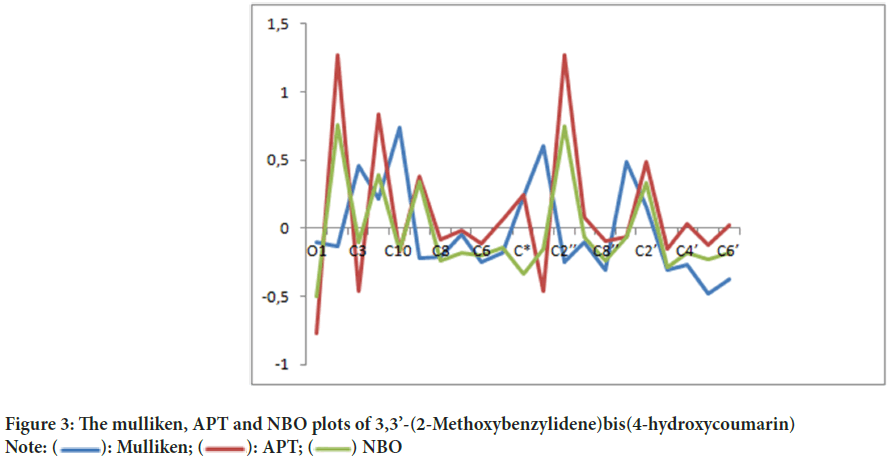
Mulliken charge distribution is an old method and common method (Mulliken RS, 1955). This method is a linear combination of atomic orbital results based on the method of obtaining molecular orbital (Figure 3). In case making the distribution to atoms of wave functions, it is based on the principle of equally distributing two overlapping orbits. Mulliken, Atomic Polar Tensor (APT), Natural Bond Orbital (NBO) charges of 3,3’-(2-Methoxybenzylidene)bis(4-hydroxycoumarin)molecule were calculated and results were given in Table 4.
| Atom | Mulliken | APT | NBO | |||
|---|---|---|---|---|---|---|
| B3LYP | HSEH1PBE | B3LYP | HSEH1PBE | B3LYP | HSEH1PBE | |
| O1 | -0,107848 | -0,058742 | -0,766021 | -0,768945 | -0,49626 | -0,48797 |
| C2 | -0,131133 | -0,173918 | 1,266719 | 1,276791 | 0,76155 | 0,76267 |
| C3 | 0,456219 | 0,457077 | -0,456636 | -0,461664 | -0,10582 | -0,12234 |
| C4 | 0,212323 | 0,286715 | 0,831514 | 0,838380 | 0,38771 | 0,36568 |
| C10 | 0,742426 | 0,837575 | -0,173008 | -0,172090 | -0,16172 | -0,07139 |
| C9 | -0,222743 | -0,275699 | 0,377594 | 0,380751 | 0,34496 | 0,31644 |
| C8 | -0,205367 | -0,256948 | -0,085227 | -0,020181 | -0,23394 | -0,24274 |
| C7 | -0,045384 | -0,087940 | -0,012467 | 0,026362 | -0,18352 | -0,19008 |
| C6 | -0,251935 | -0,303304 | -0,111337 | -0,078859 | -0,19785 | -0,20865 |
| C5 | -0,177932 | -0,211178 | 0,057813 | 0,141433 | -0,14427 | -0,17758 |
| C* | 0,233055 | 0,163381 | 0,244713 | 0,243784 | -0,33443 | -0,34663 |
| C3’’ | 0,605173 | 0,700666 | -0,458572 | -0,464806 | -0,15495 | -0,16694 |
| C2’’ | -0,246357 | -0,351988 | 1,275489 | 1,285970 | 0,75083 | 0,75242 |
| C5’’ | -0,101701 | -0,128310 | 0,076376 | 0,161992 | -0,06177 | -0,06853 |
| C8’’ | -0,304592 | -0,419762 | -0,094019 | -0,027355 | -0,23380 | -0,24170 |
| C1’ | 0,491858 | 0,594870 | -0,064988 | -0,065842 | -0,06620 | -0,07061 |
| C2’ | 0,159090 | 0,122966 | 0,489079 | 0,492519 | 0,33357 | 0,33342 |
| C3’ | -0,306666 | -0,305691 | -0,151932 | -0,110941 | -0,28596 | -0,29567 |
| C4’ | -0,267882 | -0,331063 | 0,032537 | 0,058516 | -0,18147 | -0,18669 |
| C5’ | -0,480995 | -0,538916 | -0,124592 | -0,101670 | -0,22474 | -0,23266 |
| C6’ | -0,372054 | -0,411745 | 0,025263 | 0,081983 | -0,18092 | -0,18601 |
Table 4: The mulliken, APT and NBO charge distribution calculated (with 6-311++G(d,p)) for 3,3’-(2-Methoxybenzylidene)bis(4-hydroxycoumarin) molecule
Figure 3: The mulliken, APT and NBO plots of 3,3’-(2-Methoxybenzylidene)bis(4-hydroxycoumarin)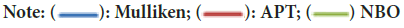
MEP surface map is very important for the investigation of molecular structure. MEP surface map shows the molecule’s color, shape, charge, size, and electrostatic potential regions at the same time. The color chart of MEP is red for (-) charge, blue for (+) charge, yellow for the slightly electron-rich region, green for neutral. 3D MEPs of the 3,3’-(2-Methoxybenzylidene) bis(4-hydroxycoumarin) molecule were simulated by using the B3LYP and shown in Figure 4.
Figure 4: MEPs maps of 3,3’-(2-Methoxybenzylidene)bis(4-hydroxycoumarin) at B3LYP/6-311G(d,p) level
Molecular docking analysis
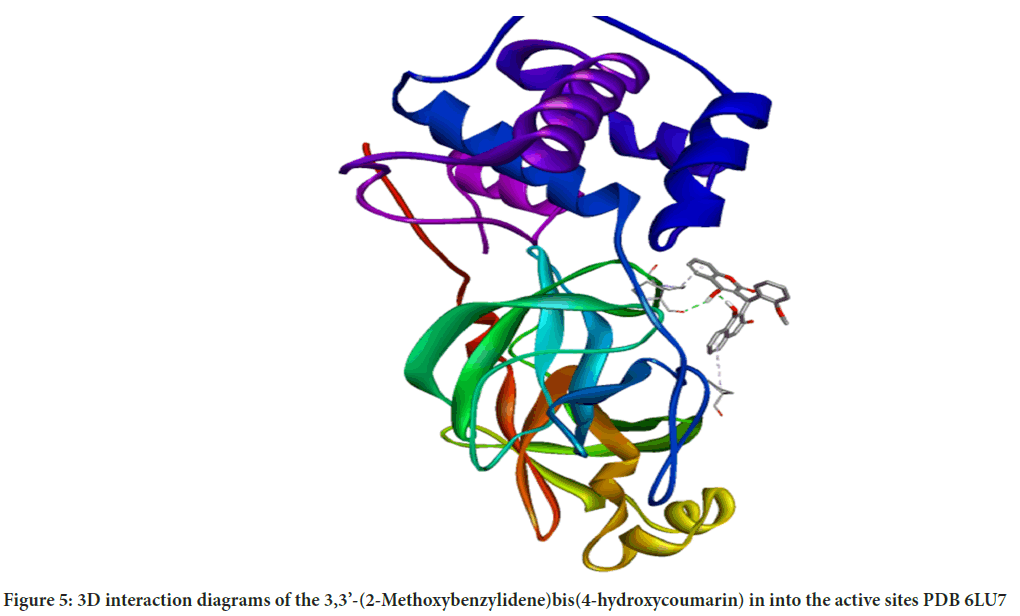
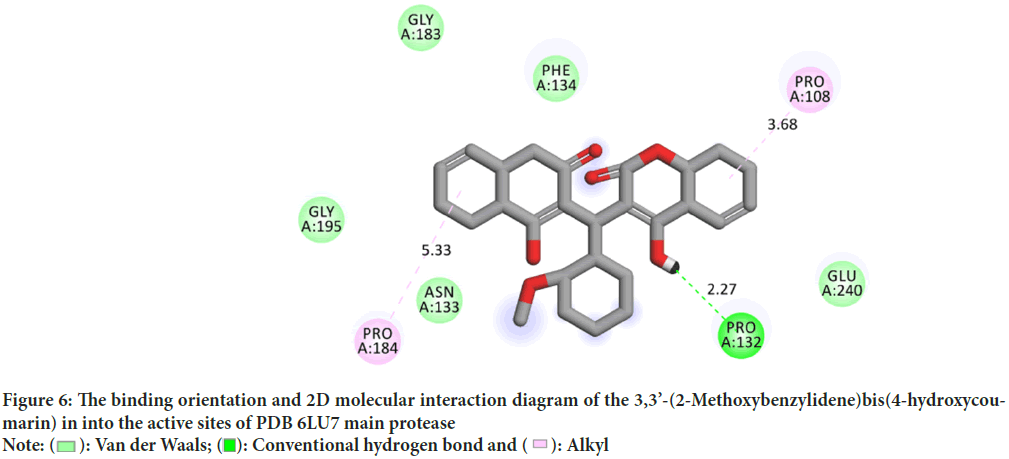
Molecular interactions play important roles in the basic biological processes. These molecular interactions lead to the formation of stable ligand-protein complexes that are necessary for their biological functions. Molecular docking was performed by AutoDock (version 4.0) program. The program receives a semi empirical free energy force in docking simulation processes. The PDB 5R7Y target protease exhibits the minimum binding energy of -4.2 kcal/mol, intermolecular energy of -4.27 kcal/mol and an inhibition constant of 836.1 uM. The deviation between the ligand-protease was analyzed, where the Root Mean Square Deviation (RMSD) value was calculated as 21.53 for 3,3’-(2-Methoxybenzylidene)bis(4-hydroxycoumarin). 3D molecular interaction diagrams of target (PDB 6LU7) and ligand (3,3’-(2-Methoxybenzylidene)bis(4-hydroxycoumarin)) are shown in Figure 5. Detailed 2D interaction diagrams are shown in Figure 6.
Figure 5: 3D interaction diagrams of the 3,3’-(2-Methoxybenzylidene)bis(4-hydroxycoumarin) in into the active sites PDB 6LU7
Figure 6: The binding orientation and 2D molecular interaction diagram of the 3,3’-(2-Methoxybenzylidene)bis(4-hydroxycoumarin) in into the active sites of PDB 6LU7 main protease
Conclusion
The geometry of 3,3’-(2-Methoxybenzylidene)bis(4-hydroxycoumarin) molecule was optimized in different levels with DFT/B3LYP and DFT/ HSEH1PBE method using 6-311G(d,p) basis set. The calculated structural parameters (bond distances, bond angles and dihedral angles) compares well with the experimental values. 3,3’-(2-Methoxybenzylidene) bis(4-hydroxycoumarin) molecule and its electrical (HOMO, HOMO-1, LUMO and LUMO+1) properties, Mulliken, NBO and Atomic Polar Tensor (APT) charges and Molecular Electrostatic Potential (MEP) surfaces have been theoretically calculated by using B3LYP (Becke’s 3-parameter hybrid functional using B exchange and LYP correlation) and HSEH1PBE (The exchange part of the screened Coulomb potential of Heyd, Scuseria, and Ernzerhof) levels of Density Functional Theory (DFT) method with 6-311++G(d,p) basis set. The active site binding properties and binding conformations were shown visually with molecular docking study.
References
- Weinmann I. History of the development and applications of coumarin and coumarin-related compounds. Coumarins: Biology, applications and mode of action. John Wiley and Sons. 1997.
- Marcu MG, Schulte TW, Neckers L. Novobiocin and related coumarins and depletion of heat shock protein 90-dependent signaling proteins. J Natl Cancer Inst. 2000; 92(3): 242-248.
[Crossref] [Google Scholar] [PubMed]
- Borges F, Roleira F, Milhazes N, Santana L, Uriarte E. Simple coumarins and analogues in medicinal chemistry: Occurrence, synthesis and biological activity. Curr Med Chem. 2005; 12(8): 887-916.
[Crossref] [Google Scholar] [PubMed]
- Chiang CC, Hsu LY, Tsai HJ, Yao CW, Chang TC. Synthesis and antimicrobial evaluation of coumarin derivatives. J Chiong Chang Inst Techol. 2008; 37: 15-22.
- Cavar S, Kovac F, Maksimovic M. Synthesis and antioxidant activity of selected 4-methylcoumarins. Food Chem. 2009; 117(1): 135-142.
- Jung JC, Park OS. Synthetic approaches and biological activities of 4-hydroxycoumarin derivatives. Molecules. 2009; 14(11): 4790-4803.
- Symeonidis T, Chamilos M, Hadjipavlou-Litina DJ, Kallitsakis M, Litinas KE. Synthesis of hydroxycoumarins and hydroxybenzo[f]- or [h]coumarins as lipid peroxidation inhibitors. Bioorg Med Chem Lett. 2009; 19(4): 1139-1142.
[Crossref] [Google Scholar] [PubMed]
- Kostova I. Studying plant-derived coumarins for their pharmacological and therapeutic properties as potential anticancer drugs. Expert Opin Drug Disc. 2007; 2(12): 1605-1618.
[Crossref] [Google Scholar] [PubMed]
- Nolan KA, Doncaster JR, Dunstan MS, Scott KA, Frenkel AD, Siegel D, et al. Synthesis and biological evaluation of coumarin-based inhibitors of NAD(P)H: Quinone Oxidoreductase-1 (NQO1). J Med Chem. 2009; 52(22): 7142-7156.
[Crossref] [Google Scholar] [PubMed]
- Mahajan DH, Pannecouque C, de Clercq E, Chikhalia KH. Synthesis and studies of new 2-(coumarin-4-yloxy)-4,6-(substituted)-s-triazine derivatives as potential anti-HIV agents. Arch Pharm. 2009; 342(5): 281-290.
[Crossref] [Google Scholar] [PubMed]
- Zhao H, Neamati N, Hong H, Mazumder A, Wang S, Sunder S, et al. Coumarin-based inhibitors of HIV integrase. J Med Chem. 1997; 40: 242-249.
[Crossref] [Google Scholar] [PubMed]
- Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Cheeseman JR, Scalmani G, et al. Gaussian 09, Revision. Wallingford CT. 2009.
- Dennington R. GaussView, version 5. 2009.
- Becke AD. Density-functional thermochemistry. V. Systematic optimization of exchange-correlation functionals. Chem Phys. 1997; 107(20): 8554-8560.
- Lee C, Yang W, Parr RG. Development of the colle-salvetti correlation-energy formula into a functional of the electron density. Phys Rev. 1988; 37(2): 785.
[Crossref] [Google Scholar] [PubMed]
- Heyd J, Scuseria GE. Efficient hybrid density functional calculations in solids: Assessment of the heyd-scuseria-ernzerhof screened coulomb hybrid functional. J Chem Phys. 2004; 121(3): 1187-1192.
[Crossref] [Google Scholar] [PubMed]
- Heyd J, Scuseria GE. Assessment and validation of a screened coulomb hybrid density functional. J Chem Phys. 2004; 120(16): 7274-7280.
[Crossref] [Google Scholar] [PubMed]
- Heyd J, Peralta JE, Scuseria GE, Martin RL. Energy band gaps and lattice parameters evaluated with the heyd-scuseria-ernzerhof screened hybrid functional. J Chem Phys. 2005; 123(17): 174101.
[Crossref] [Google Scholar] [PubMed]
- Heyd J, Scuseria GE, Ernzerhof M. Hybrid functionals based on a screened coulomb potential. J Chem Phys. 2003; 118(3): 8207-8215.
- Frisch MJ, Pople JA, Binkley JS. Self‐consistent molecular orbital methods 25. Supplementary functions for gaussian basis sets. J Chem Phys. 1984; 80(7):3265-3269.
- Novobiocin and related coumarins and depletion of heat shock protein 90-dependent signaling proteins
- Sanner MF. Python: A programming language for software integration and development. J Mol Graph Model. 1999; 17(1): 57-61.
[Google Scholar] [PubMed]
- DeLano WL. Pymol: An open-source molecular graphics tool. Protein Crystallogr. 2002; 40(1): 82-92.
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comp Chem. 2009; 30(16): 2785-2791.
[Crossref] [Google Scholar] [PubMed]
- Crossref
- Završnik D, Muratović S, Makuc D, Plavec J, Cetina M, Nagl A, et al. Benzylidene-bis-(4-hydroxycoumarin) and benzopyrano-coumarin derivatives: Synthesis, 1H/13C-NMR conformational and X-ray crystal structure studies and in vitro antiviral activity evaluations. Molecules. 2011; 16(7): 6023-6040.
[Crossref] [Google Scholar] [PubMed]
- Mulliken RS. Electronic population analysis on LCAO-MO molecular wave functions I. J Chem Phys. 1955; 23: 1833.
Author Info
Cengiz Ipek1* and Hacer Gumus22Ali Riza Veziroglu Vocational School, Kocaeli University, Kocaeli, Turkey
Citation: Ipek C, et al.: Molecular Docking Study of the Benzylidene-bis-(4-Hydroxycoumarin) Derivatives as Antiviral to Coronavirus COVID-19
Received: 12-Jan-2024 Accepted: 29-Jan-2024 Published: 05-Feb-2024, DOI: 10.31858/0975-8453.15.2.81-87
Copyright: This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
ARTICLE TOOLS
- Dental Development between Assisted Reproductive Therapy (Art) and Natural Conceived Children: A Comparative Pilot Study Norzaiti Mohd Kenali, Naimah Hasanah Mohd Fathil, Norbasyirah Bohari, Ahmad Faisal Ismail, Roszaman Ramli SRP. 2020; 11(1): 01-06 » doi: 10.5530/srp.2020.1.01
- Psychometric properties of the World Health Organization Quality of life instrument, short form: Validity in the Vietnamese healthcare context Trung Quang Vo*, Bao Tran Thuy Tran, Ngan Thuy Nguyen, Tram ThiHuyen Nguyen, Thuy Phan Chung Tran SRP. 2020; 11(1): 14-22 » doi: 10.5530/srp.2019.1.3
- A Review of Pharmacoeconomics: the key to “Healthcare for All” Hasamnis AA, Patil SS, Shaik Imam, Narendiran K SRP. 2019; 10(1): s40-s42 » doi: 10.5530/srp.2019.1s.21
- Deuterium Depleted Water as an Adjuvant in Treatment of Cancer Anton Syroeshkin, Olga Levitskaya, Elena Uspenskaya, Tatiana Pleteneva, Daria Romaykina, Daria Ermakova SRP. 2019; 10(1): 112-117 » doi: 10.5530/srp.2019.1.19
- Dental Development between Assisted Reproductive Therapy (Art) and Natural Conceived Children: A Comparative Pilot Study Norzaiti Mohd Kenali, Naimah Hasanah Mohd Fathil, Norbasyirah Bohari, Ahmad Faisal Ismail, Roszaman Ramli SRP. 2020; 11(1): 01-06 » doi: 10.5530/srp.2020.1.01
- Manilkara zapota (L.) Royen Fruit Peel: A Phytochemical and Pharmacological Review Karle Pravin P, Dhawale Shashikant C SRP. 2019; 10(1): 11-14 » doi: 0.5530/srp.2019.1.2
- Pharmacognostic and Phytopharmacological Overview on Bombax ceiba Pankaj Haribhau Chaudhary, Mukund Ganeshrao Tawar SRP. 2019; 10(1): 20-25 » doi: 10.5530/srp.2019.1.4
- A Review of Pharmacoeconomics: the key to “Healthcare for All” Hasamnis AA, Patil SS, Shaik Imam, Narendiran K SRP. 2019; 10(1): s40-s42 » doi: 10.5530/srp.2019.1s.21
- A Prospective Review on Phyto-Pharmacological Aspects of Andrographis paniculata Govindraj Akilandeswari, Arumugam Vijaya Anand, Palanisamy Sampathkumar, Puthamohan Vinayaga Moorthi, Basavaraju Preethi SRP. 2019; 10(1): 15-19 » doi: 10.5530/srp.2019.1.3