Research Article - (2022) Volume 13, Issue 4
Abstract
The papain-like protease (PLpro) is a useful target for discovering SARS-CoV-2 therapeutics. This study targets the newly emerged SARS-CoV-2 PLpro by anti-SARS-CoV PLpro, anti-HCV NS3, and anti-HIV protease drugs. Sequence analysis, modeling, and docking are used to get a valid model for SARS-CoV-2 PLpro and test the drugs' binding affinity. Results suggest the effectiveness of two anti-SARS-CoV drugs, three anti-HCV drugs, and eight antiHIV drugs as possible potent binders against the newly emerged coronavirus, SARS-CoV-2 PLpro active site. The binding affinities arise mainly from the established H-bonding and hydrophobic contacts with K105, W106, H272, and D286. The suggested compounds and drugs may be used as possible therapeutics against COVID-19.
Keywords
SARS-CoV-2, PLpro, Protease, COVID-19, Molecular docking, Drug repurposing
Introduction
A newly emerged human coronavirus (SARS-CoV-2) was reported two years ago in Wuhan, China (Hui DS, et al., 2020; Bogoch II, et al., 2020). Based on the World Health Organization (WHO) surveillance draft, any traveler to Wuhan city in Hubei Province in China 15 days before the onset of the symptoms was suspected to be a SARS-CoV-2 patient (WHO, 2020; Bogoch II, et al., 2020). WHO distributed interim guidance for laboratories that carry out tests for the emerged outbreak and released infection prevention and control guidance (WHO, 2020). SARS-CoV-2 viral pneumonia is believed to be related to the seafood market, where an unknown animal is considered responsible for the emergence of the outbreak (Hui DS, et al., 2020).
Countries other than China started borders surveillance to prevent the spread of the new coronavirus, espe en the Chinese New Year holiday w ect (Parr J, 2020). The number of infections is grossly increasing every day, and the number of confirmed cases at the time of writing this article is more than 350 million, with more than 5.6 M deaths reported worldwide (Yang L, 2020). The Nationa Commission of China confirmed the human-to-human transmission of the Wuhan outbreak (SARS-CoV-2) on January 20, 2020, (Yang L, 2020). The symptoms include fever, malaise, dry cough, shortness of breath, and respiratory distress (Hui DS, et al., 2020, Elfiky AA, 2021). SARS-CoV-2 is a member of the Betacoronaviruses family, such as the cute Respiratory Syndrome coronavirus (SARS-CoV; 774 died out of 8000 infections) and the Middle-East Respiratory Syndrome coronavirus (MERS-CoV; 858 died out of 2500 infections) (Elfiky AA, et al., 2017; Chan JF, et al., 2015). Human Coronaviruses (HCoVs) are zoonotic viruses that transmit from animals to humans through direct contact. Until today, seven different strains of HCoVs have been reported, including the newly emerged SARS-CoV-2 (WHO, 2016; Hui DS, et al., 2020). 229E and NL63 strains of HCoVs belong to Alphacoronaviruses, while OC43, HKU1, SARS-CoV, MERS-CoV, and SARS-CoV-2 belong to Betacoronaviruses (Hui DS, et al., 2020, Elfiky AA, et al., 2017). SARS-CoV has a 10% mortality rate, while MERS-CoV has a 36% mortality rate, according to the WHO (Elfiky AA, et al., 2017; WHO, 2016; Hemida MG and Alnaeem A, 2019, Santos BYM, et al., 2014; WHO, 2019). For the newly emerged coronavirus, the mortality rate is far lower (2.2%) than that of SARS-CoV and MERS-CoV, but unfortunately, it has a high transmission rate. HCoVs generally are positive-sense single RNA (30 kb) viruses. Two groups of protein characterize HCoVs; structural, such as Spike (S), Nucleocapsid (N), Matrix (M), and Envelope (E), and non-structural pr ch as RNA dependent RNA polymeras (nsp12), Chymotrypsin-like protease (3CLpro or main protease (Mpro)) and the papain-like PLpro (Elfiky AA, et m>., 2017; El nd Azzam EB, 2020). PLpro is an essential enzyme in the life cycle of RNA vir cluding the HCoVs. PLpro is a multifunctional cysteine protease that processes the viral polyprotein and host cell proteins by hydrolyzing the peptid peptide bonds in viral and cellular substrates leading to the virus replication (Santos BYM, et al., 2014). PLpro is targeted in different coronaviruses, including SARS-CoV and MERS-CoV (Elfiky AA and Ismail A, 2019; Elfiky AA, 2019; Elfiky AA and Ismail AM, 2017; Elfiky AA and Elshemey WM, 2018; Elfiky AA, 2017.). PLpro is an essential target as it is a multifunctional viral protein (Santos BYM, et al., 2015; Durai P, et al., 2015). It is responsible for the deubiquitination of IRF3, which, subsequently, inhibits Interferone β synthesis (Yang X, et al., 2014; Berman H, et al., 2003). This study generated the SARS-CoV-2 PLpro model using homology modeling after sequence comparison to the solved structures in the protein data bank (Artimo P, et al., 2012). Molecular docking is then performed to test the binding of some selected drugs (anti-SARS-CoV PLpro, anti-HCV NS3, and anti-HIV protease) against SARS-CoV-2 PLpro.
Materials and Methods
Sequence alignment and modeling
The first deposited gene for the newly emerged SARS-CoV-2 (NC_045512.2) is retrieved from the National Center for Biotechnology Information (NCBI) nucleotide database and is then translated using ExPASy translate tool (NCBI, 2020) ( , et al., 2014). The Swiss Model is used to build a SARSCoV-2 PLpro (Altschul SF, et al., 1997). Using Basic Local Alignment Search Tool (BLAST) against the SARS-CoV-2 PLpro, we found eight different solved structures for SARS-CoV PLpro (PDB IDs: 5TL6, 2FE8, 5E6J, 3MJ5, 5Y3E, 4M0W, 3E9S, and 4OVZ) that have at least 82.17% sequence ident RS-CoV-2 PLpro (SAVES, 2020). We choose the 5Y3E chain A because it has the best resolution (1.6 Å) among the eight SARS-CoV PLpro. Therefore, using the Swiss Model, 5Y3E is used as a template (82.8% identity) for building SARS-CoV-2 PLpro. Validation of the model is assessed by the Molprobity web server (Duke U ), and the Structure Analysis a cation Server (SAVES) (University of California Los Angles) (Williams CJ, et al., 2018; Laskowski RA, et al., OCHECK (Eisenberg D, et m., 1997), Verify-3D (Pontius J, et al., 1996), PROVE (Hooft RW, et al., 1996), and ERRAT (Summers KL, et al., 2012) are used to ju alidity of the model. Accordingly, the model is further energy minimized (MM3 force field) after adding missing Hydrogen atoms and removing water molecules or solvents utilizing the computational chemistry workspace SCIGRESS 3.4 and PyMol software for the protein to be ready for the docking experiments (Elfiky AA, 2019; Elfiky AA and Elshemey WM, 2018; Lii JH and Allinger NL, 1989; Elfiky AA and Ismail A, 2019; Trott O and Olson AJ, 2010).
Molecular docking
Docking experiments are performed on the optimized SARS-CoV-2 PLpro and the SARSCoV PLpro (PDB ID: 5Y3E, chain A) by the aid of AutoDock Vina software. Seventeen different compounds (three anti-SARS, three and eleven anti-HIV) are tested against SARS-CoV-2 PLpro. The used anti-SARS-CoV compo N-(1,3-benzodioxol5-ylmethyl)-1-((1R)-1-naphthalen-1-ylethyl)piperidine-4-carboxamide (GRL-0667) (NCBI, 2008), 5-amino-2-methylN-((1R)-1-naphthalen-1ylethyl) benzamide (GRL-0617) (NCBI, 2008), and mycophenolic acid (NCBI, 2005). The used anti-HCV NS3 drugs are the three approved (by the FDA) drugs, telaprevir, boceprevir (NCBI, 2006), and grazoprevir (NCBI, 2010). The anti-HIV approved drugs are amprenavir (Adkins JC and Faulds D, 1998), atazanvir (Johnson M, 1987), duranvir (Mukonzo J, et al., 2019), indinavir (Piscitelli SC, et al., 2000), leupeptin (Libby P and Goldberg AL, 1978), lopinavir (Hurst M and Faulds D, 2000), nelfinavir (Elliot BA and Plosker GL, et al., 2000), saquinavir (Noble S ds D, 1996), tiprenavir (Taura M, et al., 2013), ritonavir (Hsu A, et al 998) and tmc310911 (Dierynck I, et al., 2011; Wu C, et al., ter docking, the structures are analyzed through the Protein-Ligand Interaction Profiler (PLIP) web server (Technical University of Dresden) (Salentin S, et al., 2015).
Results and Discussion
SARS-CoV-2 PLpro modeling
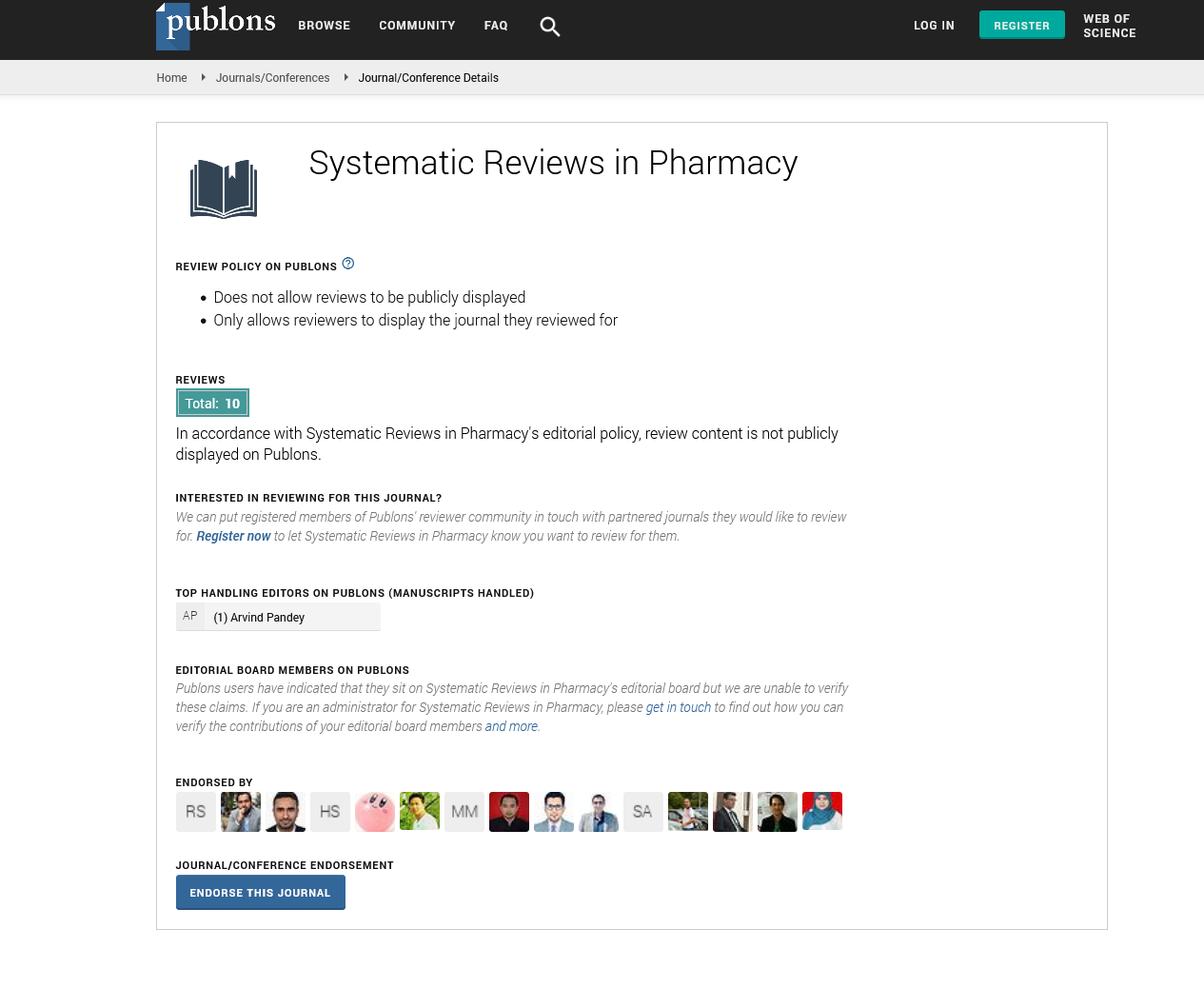
Figure1A shows the pairwise sequence alignment of the PLpro of SARS-CoV and SARSCoV-2 strains of coronavirus. SARS-CoV PLpro secondary structure is presented at the top of the alignment (PDB ID: 5Y3E chain: A), while its water accessibility is displayed at the bottom with blue indicating highly accessible residues and cyan partially accessible. At the same time, white is used for the buried residues. Three black-dashed rectangles mark the active site residues (C111, H272, and D286) of SARS-CoV and SARS-CoV-2 PLpro. Figure 1B shows the pairwise sequence alignment of SARS-CoV-2 PLpro versus HCV NS3 (PDB ID: 3SU6). Orange-da angles surround the active site residues of HCV N D102, and S159). The dashed-black rectangles mark active sites of both SARS-CoV-2 PLpro and SARS PLpro (C111, H272, and D286), while dashed-ora ngles mark the active site of HCV NS3 (H78, D102, and S159). The alignment is perform the CLUSTAL omega web server and represe Spript 3. As implied from the alignment, SARS-CoV ersus SARS-CoV-2 PLpro shows servation (highlighted in red), called sequelogous. Despite the pairwise percent identity of SARS-CoV-2 PLpro against SARS-CoV PLpro is 82.8%, and only 11.85% for HCV NS3, the sim to SARS-CoV-2 PLpro is 93.81% and 59.68% for SARS-CoV PLpro and HCV NS3, respectively. The active site triad C111, H272, and D286 of both SARS-CoV and SARS-CoV-2 PLpro (Figure 1A) are partially surface accessible in order for PLpro to be able to attack its substrates for cleavage (Malcolm B, et al., 2006). Similarly, HCV NS3 active site residues H78 and D102 are surface-affordable (Figure 1B).
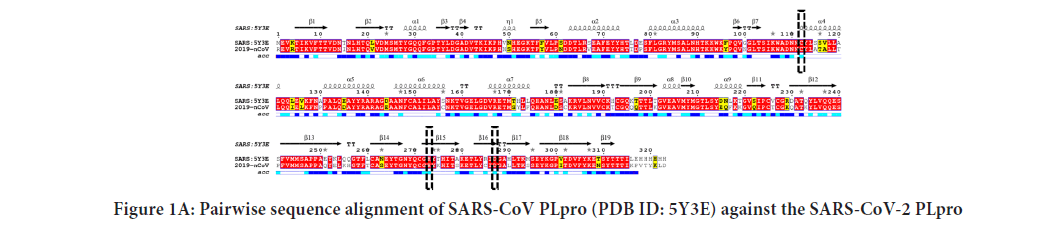
Figure 1A: Pairwise sequence alignment of SARS-CoV PLpro (PDB ID: 5Y3E) against the SARS-CoV-2 PLpro
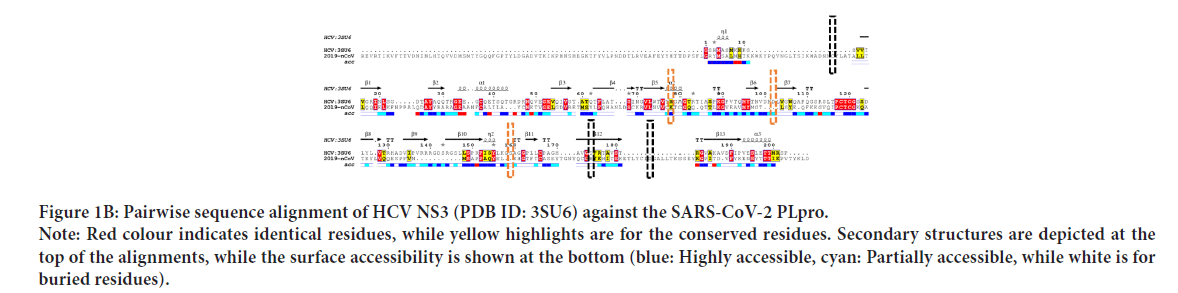
Figure 1B: 1B: Pairwise sequence alignment of HCV NS3 (PDB ID: 3SU6) against the SARS-CoV-2 PLpro. Note: Red colour indicates identical residues, while yellow highlights are for the conserved residues. Secondary structures are depicted at the top of the alignments, while the surface accessibility is shown at the bottom (blue: Highly accessible, cyan: Partially accessible, while white is for buried residues).
(Morris GM, et al., 2009). This can be deduced from Figure 1C, where the SARS-CoV-2 PLpro model is represented by PyMOL software in the surface (right) and carton (left) representations. e active site residues are in red (surface accessible), while the rest are green (see the enlarged panel). The surface accessibility is v the protease function, allowing the interaction with the substrates. The complete genome for SARS-CoV-2 has a uence identity of 89.12% and 82.34% with Bat SARS-like coronavirus isolate bat-SL-CoVZC45 and SARS coronavirus ZS-C, respectively. Drug designers should take care of the identity, especially when emerging RNA viruses that have a high mutation rate are targeted. On the other hand, potent drugs that could undetectably present on shelves can stop the rapidly developing SARS-CoV-2 strain.
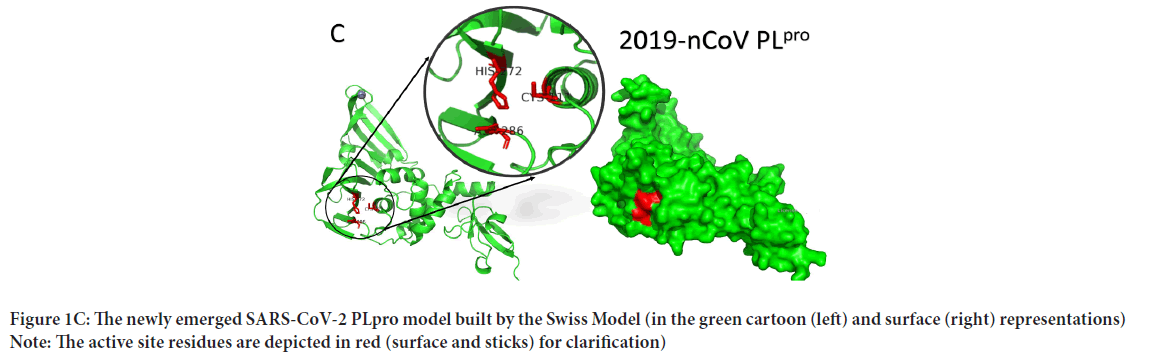
Figure 1C: The newly emerged SARS-CoV-2 PLpro model built by the Swiss Model (in the green cartoon (left) and surface (right) representations) Note: The active site residues are depicted in red (surface and sticks) for clarification)
SARS-CoV-2 PLpro model (315 residues) is built with the aid of the Swiss Model using SARS-CoV PLpro (PDB ID: 5Y3E, chain A) as a homolog. The model is sequelogous (82.8% id) to the template, reflecting the high-quality model obtained. The model is valid based on the Ramachandran plot's values (100% in the allowed region and 92.9% in the most favored region). Besides, 92.04% of the residues have averaged a 3D-1D score of ≤ 0.2 (Verify 3D software), while the overall quality factor (ERRAT software) is 94.8%. PROVE software gives 2.7% atomic volume outliers, which is acceptable.
Anti-SARS-CoV PLpro, anti-HCV NS3, and anti-HIV protease binding to SARS-CoV-2 PLpro
Before performing the docking experiments, the structures of the small molecules, the SARS-CoV-2 PLpro model, SARS-CoV PLpro structure (PDB ID: 5Y3Q, Chain A), HCV NS3 structure (PDB ID: 3SU6, Chain A), and HIV protease structure (PDB ID: 6U7O, Chain A) are prepared. The missing Hydrogen atoms are added to the protein structure and model, while any water molecules or ligands are removed. Ligand structures are retrieved from the protein data bank to be physiologically active. Figure 2 show the 2D structures of the anti-SARS-CoV PLpro, anti-HCV NS3, and anti-HIV protease compounds and drugs used in this study ((telaprevir, boceprevir, and grazoprevir), and the anti-hiv protease (amprenavir, atazanavir, duranvir, indinavir, leupeptin, lopinavir, nelfinavir, saquinavir, tipranavir, Ritonavir, and TMC310911) retrieved from the PubChem database. Carbon atoms are not explicitly represented by a letter, while N stands for Nitrogen, O stands for Oxygen, S stands for Sulfur, and H stands for Hydrogen). The anti-SARSCoV PLpro, GRL-0667, GRL-0617, and mycophenolic acids are retrieved from the PDB files 3MJ5 (GRM), 3E9S (TTT), and 1JR1 (MOA), respectively. Besides, the anti-HCV NS3; telaprevir, boceprevir, and grazoprevir are retrieved from the PDB files; 3SV6 (SV6), 3LOX (MCX), and 3SUD (SUE), respectively. Additionally, the anti-HIV protease; amprenavir, atazanavir, duranvir, indinavir, leupeptin, lopinavir, nelfinavir, saquinavir, tipranavir, ritonavir, and TMC310911, are retrieved from the PDB files; 3OXV, 3OXX, 3OXW, 3WSJ, 6BKJ, 6DJ1, 3EKX, 4Q5M, 6DIF, 5VC0, and 3R4B, respectively.
Figure 2: 2D structures of the anti-SARS-CoV PLpro (top) (GRL-0667, GRL-0617, and My c acid), anti-HCV NS3 drugs
The active site of the proteins is treated as flexible during all the docking experiments, while the exhaustiveness is unified at 8. A grid box of size 30 Å × 36 Å × 30 Å centered at (-17.9, 43.9, 1.6) Å is prepared for SARSCoV-2 PLpro utilizing the AutoDock tools (61). Additionally, grid boxes of almost identical sizes centered at the active site residues are made for SARS-CoV PLpro, HCV NS3, and HIV protease. AutoDock Vina is used to predict the interaction between the anti-SARS-CoV, anti-HCV NS3, and anti-HIV protease drugs and the active site of SARS-CoV-2 PLpro. Figure 3A shows the binding affinities (docking scores in kcal/mol) for the docking of the anti-SARS-CoV and anti-HCV drugs against SARS-CoV-2 PLpro (blue), SARS-CoV PLpro (orange), and HCV NS3 (gray). Besides, Figure 3B shows the binding affinities for the docking of anti-HIV drugs against SARS-CoV-2 PLpro (blue), SARSCoV PLpro (orange), and HIV protease (gray). The binding energies for SARS-CoV-2 PLpro are listed for each ligand in Figures3A and 3B.
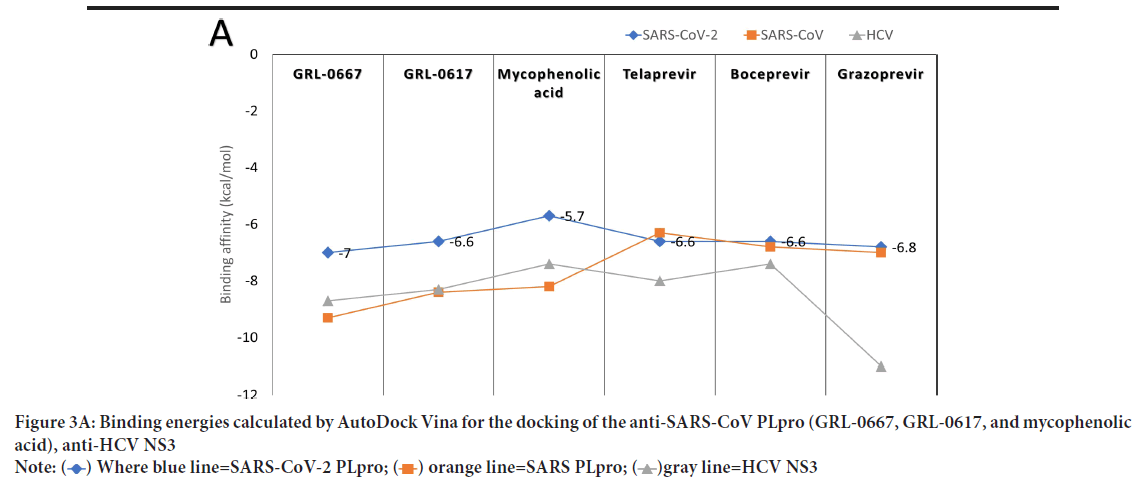
Figure 3A: Binding energies calculated by AutoDock Vina for the docking of the anti-SARS-CoV PLpro (GRL-0667, GRL-0617, and mycophenolic
acid), anti-HCV NS3
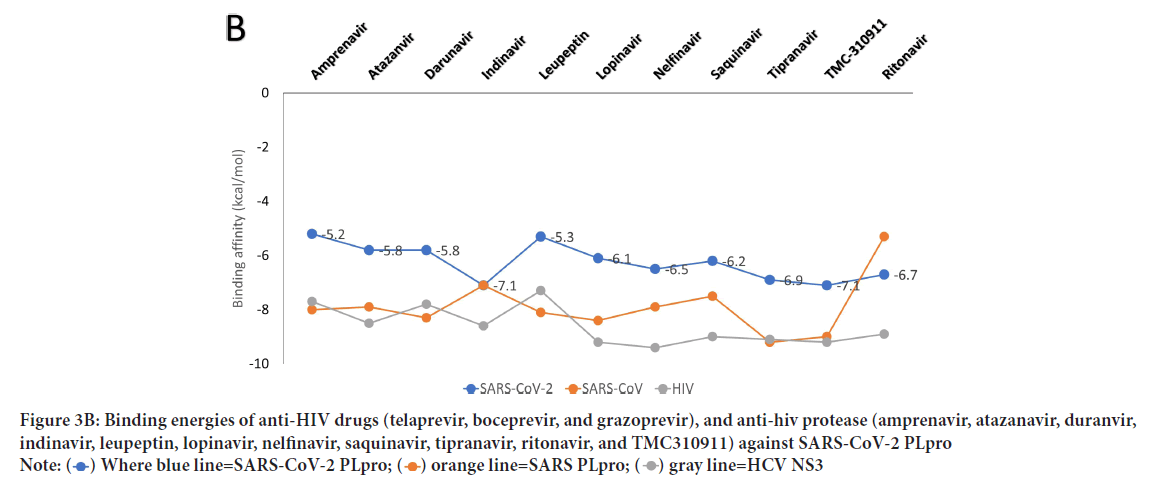
Figure 3B: Binding energies of anti-HIV drugs (telaprevir, boceprevir, and grazoprevir), and anti-hiv protease (amprenavir, atazanavir, duranvir, indinavir, leupeptin, lopinavir, nelfinavir, saquinavir, tipranavir, ritonavir, and TMC310911) against SARS-CoV-2 PLpro 
As reflected from the docking scores in Figure 3A, six compounds can bind to SARSCoV-2 PLpro, SARS-CoV PLpro, and HCV NS3 with variable binding energy values (-5.7 up to11.0 kcal/mol). Compared to other proteases, the binding energies for the drugs to SARS-CoV-2 PLpro are less negative (lower affinity). For GRL-0667, GRL-0617, and Mycophenolic acid, the reductions in the binding energies for SARS-CoV-2 PLpro are 25%, 21%, and 30%, respectively, compared to SARS-CoV PLpro. In HCV, the decreases in the SARS-CoV-2 PLpro binding energies are 18%, 11%, and 38% for telaprevir, boceprevir, and grazoprevir, respectively. Despite these reductions, the PLpro of SARS-CoV-2 can still bind to the drugs with good binding energies (-5.7 up to -7 kcal/mol), which is enough to maintain the viral protein dysfunctionally. For the anti-HIV drugs, the best four compounds that can bind to SARS-CoV-2 PLpro active site are indinavir (-7.1 kcal/mol), TMC310911 (-7.1 kcal/mol), tiprenavir (-6.9 kcal/ mol), and ritonavir (-6.7 kcal/mol). Other anti-HIV drugs give moderate binding energies to SARS-CoV-2 PLpro (higher than -6.5 kcal/mol) except for amprenavir and leupeptin, which provide lower binding affinities (-5.2 and -5.3 kcal/mol, respectively). The anti-HIV drugs can bind to both HIV protease and SARS-CoV PLpro with almost the same binding affinity (-7.1 and down to -9.4 kcal/mol) with ritonavir as an exception (-5.3 kcal/mol for SARS-CoV PLpro).
To further analyze the binding patterns, we examined the i n complexes formed upon docking by the PLIP web server's aid. Figures 4 A-C show the interactions formed after docking of the anti-SARS-CoV (GRL0667, GRL-0617, and Mycophenolic acid), anti-HCV drugs (telaprevir, boceprevir, and grazoprevir), and anti-HIV protease drugs (Indinavir, TMC310911, Tiprenavir, and Ritonavir), respectively. The solid blue lines indicate H-bonding, while dashed grey lines indicate hydrophobic interactions. The salt bridges and the π-π contacts are represented in balls connected by yellow dashed lines and green dashed lines. The labeled residues (blue sticks) represent the SARS-CoV-2 active residues that interact with the ligands (orange sticks). The details of the interactions are tabulated in Tables 1-3for the anti SARS-CoV, anti-HCV, and anti-HIV drugs, respectively. The binding energies are listed in the table, along with the number of formed H-bonds and the number of hydrophobic contacts established after docking. The amino acids from the SARS-CoV PLpro, SARS-CoV-2 PLpro, HCV NS3, and HIV protease that interact with the ligands are also listed in the tables. The most-reported residues from the SARS-CoV-2 PLpro that interact through H-bonding or hydrophobic interactions are K105, W106, H272, and D286 (bold residues in Tables1-3). As implied from the tables, SARS-CoV PLpro, HCV NS3, and HIV protease reveal more established interactions upon docking the tested drugs than the new coronavirus strain PLpro. However, a minimum number of established interactions (five interactions) stabilize the drugs in the protein active site for SARS-CoV-2 PLpro. Besides, in all docking experiments, the established interactions are both H-bonding and hydrophobic contacts in addition to a few salt bridges (stared residues in Tables1 and 2), suggesting the possibility that these drugs (anti-SARS, anti-HCV, and anti-HIV drugs) could bind to and inhibit the function of the crucial viral enzyme PLpro of SARS-CoV-2. In summary, the present results suggest possible inhibition of some of the currently available therapeutics against the newly emerged coronavirus in silico. Anti-SARS-CoV PL pro (G nd GRL-0617), anti-HCV NS3 (telaprevir, boceprevir, and grazoprevir), and antiHIV protease drugs (indinavir, lopinavir, nelfinavir, saquinavir, tipranavir, ritonavir, and TMC310911) may bind to the active site (C112, H273, and D287) of SARS-CoV-2 PLpro. The calculated binding energies for the tested drugs against SARS-CoV-2 PLpro are slightly higher (lower affinity) than that of SARS-CoV PLpro, HCV NS3, and HIV protease. On the other hand, at least five interactions are established between SARS-CoV-2 PLpro and each tested compound (either H-bonds or hydrophobic interactions), suggesting possible targeting of SARS-CoV-2 PLpro using anti-SARS, anti-HCV NS3, and anti-HIV protease drugs. v class="table-responsive">
| Ligand (Anti-SARS) | Target | Docking score (kcal/mol) | H-bonding | Hydrophobic interaction | ||
|---|---|---|---|---|---|---|
| Number | Residues involved | Number | Residues involved | |||
| GRL-0667 | SARS-CoV-2 | -7.0 | 3 | W93, K105, W106 | 7 | K92, W106(2), W106**(3), A107 |
| SARS-CoV | -9.3 | 6 | W107, C112, G164, D165*, G272, Y274 | 5 | Y113, L163, Y265, Y274(2) | |
| GRL-0617 | SARS-CoV-2 | -6.6 | 2 | L162, Y264 | 8 | L162, P247, P248, Y264(2), Y268(2), T301 |
| SARS-CoV | -8.4 | 4 | L163, D165(2), Y274 | 5 | D165, P249, Y265, Y269, T302 | |
| Mycophenolic Acid | SARS-CoV-2 | -5.7 | 7 | K105*, W106 (2), H272, H272*, D286, A288 | 1 | L289 |
| SARS-CoV | -8.2 | 3 | G164, G272, Y274 | 1 | Y265 | |
Note: Where (*) represents salt bridges, while (**) represent π-π contact
Table 1: The interactions formed between anti-SARS compounds (GRL-0667, GRL-0617, and Mycophenolic acid) and SARS-CoV-2 PLpro upon docking
| Ligand (Anti-HCV) | Target | Docking score (kcal/mol) | H-bonding | Hydrophobic interaction | ||
|---|---|---|---|---|---|---|
| Number | Residues involved | Number | Residues involved | |||
| Telaprevir | SARS-CoV-2 | -6.6 | 8 | W106, T265, H272, K274, D286(3), A288 | 3 | K105, W106(2) |
| HCV | -8.0 | 10 | Q59(3), H75, K154, G155, S157(2), A175(2) | 6 | I150, L153, K154, F172, A174(2) | |
| Boceprevir | SARS-CoV-2 | -6.6 | 4 | W106, N109(2), H272 | 5 | K105, W106(4) |
| HCV | -7.4 | 10 | H75(2), G155, S157(3), R173, | 7 | H75(2), I150, K154(2), V176, D186 | |
| Grazoprevir | SARS-CoV-2 | -6.8 | 4 | K105, W106, K274, D286 | 3 | W106, T265, L289 |
| HCV | -11.0 | 10 | H75(2), G155, S156, S157(3), R173, A175(2) | 13 | Q59, F61(2), H75(2), H75**, D99, I150, F172, R173, A175, V176, D186 | |
Note: (*) represents salt bridges, while (**) represent π-π contact
Table 2: The interactions formed between anti-HCV compounds (telaprevir, boceprevir, and grazoprevir) and SARS-CoV-2 PLpro upon docking
| Ligand (Anti-HIV) | Target | Docking score (kcal/mol) | H-bonding | Hydrophobic interaction | ||
|---|---|---|---|---|---|---|
| Number | Residues involved | Number | Residues involved | |||
| Amprenavir | SARS-CoV-2 | -5.2 | 3 | Q269, K274, D286 | 2 | T265, L289 |
| HIV | -7.7 | 10 | L23, V32, P81, V82 | 6 | I50(2) | |
| SARS-CoV | -8 | 5 | A247, Y274(2), G272, D303 | 5 | R167, P249, Y265(2), T302 | |
| Ataznvir | SARS-CoV-2 | -5.8 | 5 | W106, N109(3), H272 | 4 | W106(2), H272, L289 |
| HIV | -8.5 | 7 | D25(2), G27, D29, D30, 48, I50 | 6 | V32, I47(3), I54, I84 | |
| SARS-CoV | -7.9 | 6 | N110(2), H273, R285(2), H290 | 8 | W107(2), N110(2), C112, L163, Y274(2) | |
| Darunavir | SARS-CoV-2 | -5.8 | 2 | W106, A288 | 3 | T265, H272, L289 |
| HIV | -7.8 | 4 | I50(2), T80 | 6 | V32, I47(2), I54, P81, V82 | |
| SARS-CoV | -8.3 | 4 | D165, G272, T302 | 4 | L163, P249, Y265, T269 | |
| Indinavir | SARS-CoV-2 | -7.1 | 3 | Q122, R140, K279 | 4 | Q121, R140, K279(2) |
| HIV | -8.6 | 4 | G48(2), G49, I50 | 4 | L23, A28, I47, V82 | |
| SARS-CoV | -7.1 | 4 | D273(3) | 7 | K106(2), W107(3), A108, D287 | |
| Leupeptin | SARS-CoV-2 | -5.3 | 5 | W106, N109(2), H272, K274 | 2 | H272, L357 |
| HIV | -7 | 5 | G49, I50, G52, I54, L357 | 3 | I54(2), L357 | |
| SARS-CoV | -8.1 | 6 | D165, R167, Y274(2), T302(2) | 1 | L357 | |
| Lopinavir | SARS-CoV-2 | -6.1 | 5 | C270, H272, D 286(3) | 7 | K105, W106(2), T265, H272, A288, L289 |
| HIV | -9.2 | 2 | I50 (2) | 8 | L23, V32, I47, 54, P81, V82 (2) | |
| SARS-CoV | -8.1 | 2 | Y269 (2) | 7 | L163 (2), D165, P249, Y265 (2), T302 | |
| Nelfinavir | SARS-CoV-2 | -6.5 | 3 | K105, W106(2) | 5 | K105, W106, A288, L289(2) |
| HIV | 8 | 8 | I3, L24, T226(2), G94, T96(3) | 13 | P1, Q2, I3, L5(2), L24, L90(2), I93, A95(2), T96, L97 | |
| SARS-CoV | -7.9 | 1 | D165 | 8 | L163, R167, P249, Y265(2), Y269, Y274, T302 | |
| Saquinavir | SARS-CoV-2 | -6.2 | 1 | H272 | 4 | W106(3), A288 |
| HIV | -9 | 5 | G49, I50(3), T80 | 10 | A28, D29, V32, I47, I50(2), I54, P81, V82, I84 | |
| SARS-CoV | -7.5 | 3 | H273, D287, H290 | 7 | K106(2), W107(2), C112, H273, H290 | |
| Tipranavir | SARS-CoV-2 | -6.9 | 3 | W106, G271, H272 | 3 | W106, H272, L289 |
| HIV | -9.1 | 4 | D25, I50(2), T80 | 9 | V32, I47(2), I54(3), P81, V82, I84 | |
| SARS-CoV | -9.2 | 1 | Y265 | 6 | L163, P249, Y265, N268, Y269, T302 | |
| TMC310911 | SARS-CoV-2 | -7.1 | 2 | R140, K279 | 3 | Y136, K279, Y283 |
| HIV | -9.2 | 2 | I50(2) | 6 | L5, V32(2), I47, P81, V82 | |
| SARS-CoV | -9 | 5 | D165, Y265(3), G272 | 7 | L163, R167, P248, P249, Y274, P300, T302 | |
| Ritonavir | SARS-CoV-2 | -6.7 | 7 | K105, W106(2), H272, K274, D286(2) | 7 | K92, W106(2), A288, L289(3) |
| HIV | -8.9 | 10 | P1, I3(2), L5, T96(3), N98(3) | 14 | P1, Q2, T4, L24(2), T26, L90(2), I93, A95(2), L97, F99 | |
| SARS-CoV | -5.3 | 6 | R285, T292, K293, S295, E296(2) | 2 | T292, Y297 | |
Table 3: The interactions formed between anti-HIV protease drugs (amprenavir, atazanavir, duranvir, indinavir, leupeptin, lopinavir, nelfinavir, saquinavir, tipranavir, ritonavir, and TMC310911) and SARS-CoV-2 PLpro upon docking
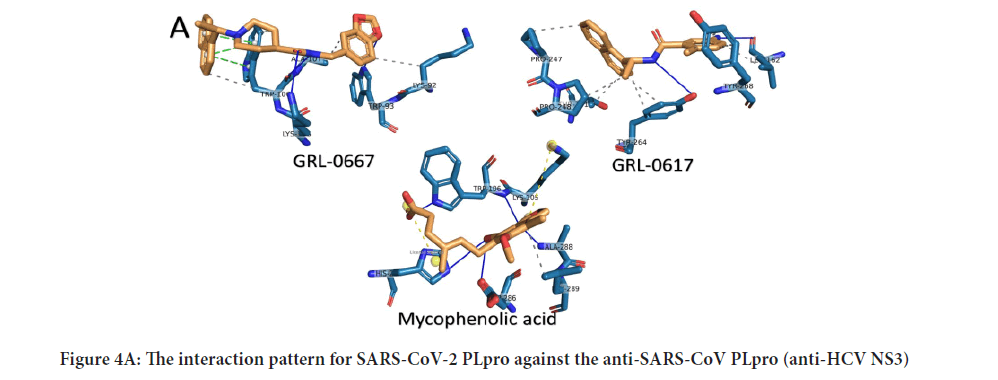
Figure 4A: The interaction pattern for SARS-CoV-2 PLpro against the anti-SARS-CoV PLpro (anti-HCV NS3)
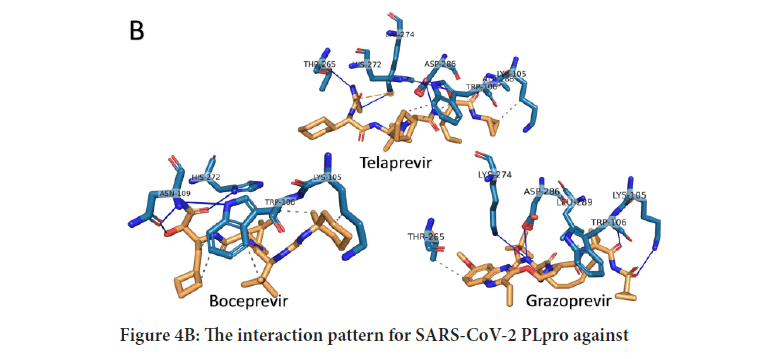
Figure 4B: The interaction pattern for SARS-CoV-2 PLpro against
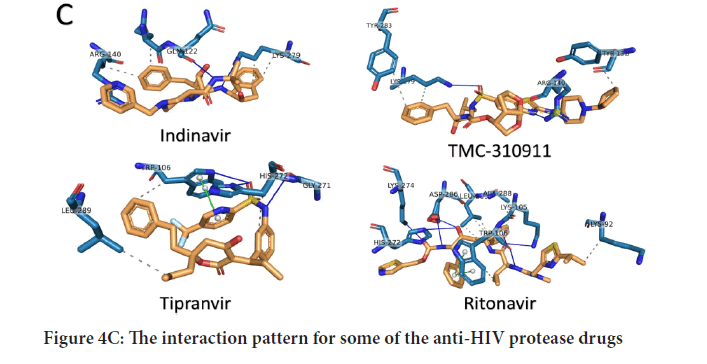
Figure 4C: The interaction pattern for some of the anti-HIV protease drugs
Conclusion
SARS-CoV-2, the causative agent of the COVID-19 pandemic, represents a significant health concern due to the vastly growing number of infections and mortalities. The present study aims to test and suggest possible inhibitor drugs for use against SARS-CoV PLpro. Anti-SARS PLpro (GRL-0667 and GRL-0617), anti-HCV NS3 (Telaprevir, boceprevir, and grazoprevir), and anti-HIV protease drugs (indinavir, lopinavir, nelfinavir, saquinavir, tipranavir, ritonavir, and TMC310911) show good binding affinities to the active site of SARS-CoV-2 PLpro and hence, may oppose viral replication. These compounds could be tested in vitrofor their effectiveness as antiSARS-CoV-2 PLpro inhibitors. Additionally, it can be used as a seed for more potent inhibitors against SARS-CoV-2 PLpro.
Acknowledgment
Noha Samir is acknowledged for her help with some calculations.
Author Contributions
A.E. has drafted the manuscript and prepared the figures and tables.
References
- Hui DS, I Azhar E, Madani TA, Ntoumi F, Kock R, Dar O, et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health-the latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. 2020; 91: 264-266.
[Crossref] [Google scholar] [Pubmed]
- Bogoch II, Watts A, Bachli AT, Huber C, Kraemer MUG, Khan K. Pneumonia of unknown etiology in Wuhan, China: Potential for international spread via commercial air travel. J Travel Med. 2020; 27(2): taaa008.
[Crossref] [Google scholar] [Pubmed]
- Surveillance case definitions for human infection with novel coronavirus (nCoV): Interim guidance v1, January 2020. World Health Organization (WHO). 2020.
- Laboratory testing of human suspected cases of novel coronavirus (nCoV) infection: Interim guidance, January 10 2020. World Health Organization (WHO). 2020.
- Infection prevention and control during health care when novel coronavirus (nCoV) infection is suspected: Interim guidance, January 2020. World Health Organization (WHO). 2020.
- Parr J. Pneumonia in China: Lack of information raises concerns among Hong Kong health workers. BMJ. 2020; 368: m56.
[Crossref] [Google scholar] [Pubmed]
- Yang L. China confirms human-to-human transmission of coronavirus. The guardian. 2020.
- Elfiky AA. SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: An in silico perspective. J Biomol Struct Dyn. 2021; 39(9): 1-9.
[Crossref] [Google scholar] [Pubmed]
- Elfiky AA, Mahdy SM, Elshemey WM. Quantitative structure-activity relationship and molecular docking revealed a potency of anti-hepatitis C virus drugs against human corona viruses. J Med Virol. 2017; 89(6): 1040-1047.
[Crossref] [Google scholar] [Pubmed]
- Chan JF, Lau SK, To KK, Cheng VC, Woo PC, Yuen KY. Middle East respiratory syndrome coronavirus: Another zoonotic betacoronavirus causing SARS-like disease. Clinical microbiol rev. 2015; 28(2): 465-522.
[Crossref] [Google scholar] [Pubmed]
- WHO. Middle East respiratory syndrome coronavirus (MERS-CoV). WHO. 2016.
- Hemida MG, Alnaeem A. Some One health based control strategies for the Middle East respiratory syndrome coronavirus. One Health. 2019; 8: 100102. [Crossref]
[Google scholar] [Pubmed]
- Santos BYM, Mielech AM, Deng X, Baker S, Mesecar AD. Catalytic function and substrate specificity of the papain-like protease domain of Nsp3 from the Middle East respiratory syndrome coronavirus. Journal of Virol. 2014; 88(21): 12511-12527.
[Crossref] [Google scholar] [Pubmed]
- Clinical management of severe acute respiratory infection when Middle East respiratory syndrome coronavirus (MERS-CoV) infection is suspected: Interim guidance. World Health Organization (WHO). 2019.
- Elfiky AA, Azzam EB. Novel guanosine derivatives against MERS CoV polymerase: An in silico perspective. J Biomol Struct Dyn. 2020; 39(8): 1-9.
[Crossref] [Google scholar] [Pubmed]
- Elfiky AA, Ismail A. Molecular dynamics and docking reveal the potency of novel GTP derivatives against RNA dependent RNA polymerase of genotype 4a HCV. Life Sci. 2019. 238: 116958.
[Crossref] [Google scholar] [Pubmed]
- Elfiky AA. Novel guanosine derivatives as anti-hcv ns5b polymerase: A QSAR and molecular docking study. Medicinal Chem. 2019; 15(2): 130-137.
- Elfiky AA, Ismail AM. Molecular modeling and docking revealed superiority of IDX-184 as HCV polymerase inhibitor. Future Virol. 2017; 12(7): 339-347.
- Elfiky AA, Elshemey WM. Molecular dynamics simulation revealed binding of nucleotide inhibitors to ZIKV polymerase over 444 nanoseconds. J Med Virol. 2018; 90(1): 13-18.
[Crossref] [Google scholar] [Pubmed]
- Elfiky AA. Zika virus: Novel guanosine derivatives revealed strong binding and possible inhibition of the polymerase. Future Virol. 2017; 12(12): 721-728.
[Crossref] [Google scholar] [Pubmed]
- Santos BYM, John SES, Mesecar AD. The SARS-coronavirus papain-like protease: Structure, function and inhibition by designed antiviral compounds. Antiviral res. 2015; 115: 21-38.
[Crossref] [Google scholar] [Pubmed]
- Durai P, Batool M, Shah M, Choi S. Middle East respiratory syndrome coronavirus: Transmission, virology and therapeutic targeting to aid in outbreak control. Exp Mol Med. 2015; 47(8): e181-e181. Crossref]
- Yang X, Chen X, Bian G, Tu J, Xing Y, Wang Y, Chen Z. Proteolytic processing, deubiquitinase and interferon antagonist activities of middle East respiratory syndrome coronavirus papain-like protease. J Gen Virol. 2014; 95(Pt 3): 614-626.
[Crossref] [Google scholar] [Pubmed]
- Berman H, Henrick K, Nakamura H. Announcing the worldwide protein data bank. Nat Struct Mol Biol. 2003; 10(12): 980-980.
[Crossref] [Google scholar] [Pubmed]
- Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, et al. ExPASy: SIB bioinformatics resource portal. Nucleic acids research. 2012; 40(W1): W597-W603.
[Crossref] [Google scholar] [Pubmed]
- National Center of Biotechnology Informatics (NCBI) database website. 2020.
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014; 42(W1): W252-W258.
[Crossref] [Google scholar] [Pubmed]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic acids research. 1997; 25(17): 3389-3402.
[Crossref] [Google scholar] [Pubmed]
- Structural analysis and verification server website (SAVES). 2020.
- Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, et al. MolProbity: More and better reference data for improved all‐atom structure validation. Protein Sci. 2018; 27(1): 293-315
[Crossref] [Google scholar] [Pubmed]
- Laskowski RA, Rullmann JAC, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996; 8(4): 477-486.
- Eisenberg D, Lüthy R, Bowie JU. VERIFY3D: Assessment of protein models with three-dimensional profiles. Methods enzymol. 1997; 277: 396-404
[Crossref] [Google scholar] [Pubmed]
- Pontius J, Richelle J, Wodak SJ. Deviations from standard atomic volumes as a quality measure for protein crystal structures. J mol biol. 1996; 264: 121-136.
[Crossref] [Google scholar] [Pubmed]
- Hooft RW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996; 381(6580): 272.
[Crossref] [Google scholar] [Pubmed]
- Summers KL, Mahrok AK, Dryden MD, Stillman MJ. Structural properties of metal-free apometallothioneins. Biochem Biophys Res Commun. 2012; 425(2): 485-492.
- Elfiky AA. The antiviral sofosbuvir against mucormycosis: An in silico perspective. Future Virol. 2019; 14(11): 739-744. [Crossref]
- Lii JH, Allinger NL. Molecular mechanics. The MM3 force field for hydrocarbons. 3. The van der Waals' potentials and crystal data for aliphatic and aromatic hydrocarbons. J Am Chem Soc. 1989; 111 (23): 8576-8582.
- Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010; 31(2): 455-461.
[Crossref] [Google scholar] [Pubmed]
- GRL0617 (CID=24941262). PubChem Database (NCBI). 2008.
- Mycophenolic acid (CID=446541). PubChem Database (NCBI). 2005.
- Telaprevir (CID=3010818). PubChem Database (NCBI). 2005.
- Boceprevir (CID=10324367). PubChem Database (NCBI). 2006.
- Grazoprevir (CID=44603531). PubChem Database (NCBI). 2010.
- Adkins JC, Faulds D. Amprenavir. Drugs. 1998; 55(6): 837-842.
[Crossref] [Google scholar] [Pubmed]
- Johnson M. Response to ‘Atazanvir/ritonavir versus lopinavir/ritonavir: Equivalent or different efficacy profiles?’by Hill. Aids. 2006; 20(15): 1987.
[Crossref] [Google scholar] [Pubmed]
- Mukonzo J, Aklillu E, Marconi V, Schinazi RF. Potential drug-drug interactions between antiretroviral therapy and treatment regimens for multi-drug resistant tuberculosis: Implications for HIV care of MDR-TB co-infected individuals. Int J Infect Dis. 2019; 83: 98-101.
[Crossref] [Google scholar] [Pubmed]
- Piscitelli SC, Burstein AH, Chaitt D, Alfaro RM, Falloon J. Indinavir concentrations and St John's wort. The Lancet. 2000; 355(9203): 547-548.
[Crossref] [Google scholar] [Pubmed]
- Libby P, Goldberg AL. Leupeptin, a protease inhibitor, decreases protein degradation in normal and diseased muscles. Science. 1978; 199(4328): 534-536.
[Crossref] [Google scholar] [Pubmed]
- Hurst M, Faulds D. Lopinavir. Drugs. 2000; 60(6): 1371-1379.
[Crossref] [Google scholar] [Pubmed]
- Elliot BA, Plosker GL. Nelfinavir. Drugs. 2000; 59(3): 581-620.
[Crossref] [Google scholar] [Pubmed]
- Noble S, Faulds D. Saquinavir. Drugs. 1996; 52(1): 93-112.
[Crossref] [Google scholar] [Pubmed]
- Taura M, Kariya R, Kudo E, Goto H, Iwawaki T, Amano M, et al. Comparative analysis of ER stress response into HIV protease inhibitors: Lopinavir but not darunavir induces potent ER stress response via ROS/JNK pathway. Free Radic Biol Med. 2013; 65: 778-788.
[Crossref] [Google scholar] [Pubmed]
- Hsu A, Granneman GR, Bertz RJ. Ritonavir. Clinical pharmacokinetics and interactions with other anti-HIV agents. Clin Pharmacokinet. 1998; 35(4): 275-291
[Crossref] [Google scholar] [Pubmed]
- Dierynck I, van Marck H, van Ginderen M, Jonckers TH, Nalam MN, Schiffer CA, et al. TMC310911, a novel human immunodeficiency virus type 1 protease inhibitor, shows in vitro an improved resistance profile and higher genetic barrier to resistance compared with current protease inhibitors. Antimicrob Agents Chemother. 2011; 55(12): 5723-5731.
[Crossref] [Google scholar] [Pubmed]
- Wu C, Liu Y, Yang Y, Zhang P, Zhong W, Wang Y, et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020; 10(5): 766-788.
[Crossref] [Google scholar] [Pubmed]
- Salentin S, Schreiber S, Haupt VJ, Adasme MF, Schroeder M. PLIP: Fully automated protein-ligand interaction profiler. Nucleic acids res. 2015; 43(W1): W443-W447
[Crossref] [Google scholar] [Pubmed]
- Malcolm B, Liu R, Lahser F, Agrawal S, Belanger B, Butkiewicz N, et al. SCH 503034, a mechanism-based inhibitor of hepatitis C virus NS3 protease, suppresses polyprotein maturation and enhances the antiviral activity of alpha interferon in replicon cells. Antimicrob Agents Chemother. 2006; 50(3):1013-1020
[Crossref] [Google scholar] [Pubmed]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009; 30(16): 2785-2791.
[Crossref] [Google scholar] [Pubmed]
Author Info
Abdo A Elfiky*Citation: Elfiky AA: SARS-CoV-2 Papain-like Protease as a Target for Anti-HCV and Anti-HIV Proteases: In silico Perspective
Received: 08-Mar-2022 Accepted: 29-Mar-2022 Published: 05-Apr-2022, DOI: 10.31858/0975-8453.13.4.242-250
Copyright: This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
ARTICLE TOOLS
- Dental Development between Assisted Reproductive Therapy (Art) and Natural Conceived Children: A Comparative Pilot Study Norzaiti Mohd Kenali, Naimah Hasanah Mohd Fathil, Norbasyirah Bohari, Ahmad Faisal Ismail, Roszaman Ramli SRP. 2020; 11(1): 01-06 » doi: 10.5530/srp.2020.1.01
- Psychometric properties of the World Health Organization Quality of life instrument, short form: Validity in the Vietnamese healthcare context Trung Quang Vo*, Bao Tran Thuy Tran, Ngan Thuy Nguyen, Tram ThiHuyen Nguyen, Thuy Phan Chung Tran SRP. 2020; 11(1): 14-22 » doi: 10.5530/srp.2019.1.3
- A Review of Pharmacoeconomics: the key to “Healthcare for All” Hasamnis AA, Patil SS, Shaik Imam, Narendiran K SRP. 2019; 10(1): s40-s42 » doi: 10.5530/srp.2019.1s.21
- Deuterium Depleted Water as an Adjuvant in Treatment of Cancer Anton Syroeshkin, Olga Levitskaya, Elena Uspenskaya, Tatiana Pleteneva, Daria Romaykina, Daria Ermakova SRP. 2019; 10(1): 112-117 » doi: 10.5530/srp.2019.1.19
- Dental Development between Assisted Reproductive Therapy (Art) and Natural Conceived Children: A Comparative Pilot Study Norzaiti Mohd Kenali, Naimah Hasanah Mohd Fathil, Norbasyirah Bohari, Ahmad Faisal Ismail, Roszaman Ramli SRP. 2020; 11(1): 01-06 » doi: 10.5530/srp.2020.1.01
- Manilkara zapota (L.) Royen Fruit Peel: A Phytochemical and Pharmacological Review Karle Pravin P, Dhawale Shashikant C SRP. 2019; 10(1): 11-14 » doi: 0.5530/srp.2019.1.2
- Pharmacognostic and Phytopharmacological Overview on Bombax ceiba Pankaj Haribhau Chaudhary, Mukund Ganeshrao Tawar SRP. 2019; 10(1): 20-25 » doi: 10.5530/srp.2019.1.4
- A Review of Pharmacoeconomics: the key to “Healthcare for All” Hasamnis AA, Patil SS, Shaik Imam, Narendiran K SRP. 2019; 10(1): s40-s42 » doi: 10.5530/srp.2019.1s.21
- A Prospective Review on Phyto-Pharmacological Aspects of Andrographis paniculata Govindraj Akilandeswari, Arumugam Vijaya Anand, Palanisamy Sampathkumar, Puthamohan Vinayaga Moorthi, Basavaraju Preethi SRP. 2019; 10(1): 15-19 » doi: 10.5530/srp.2019.1.3