Research Article - (2023) Volume 14, Issue 11
Abstract
Leukemia, a type of cancer that affects the blood and bone marrow, is a complex and devastating disease with limited treatment options. Over the years, researchers have been exploring novel therapeutic approaches to improve patient outcomes. One emerging area of interest is the role of exosomes in leukemia. Exosomes are small extracellular vesicles secreted by various cell types, including cancer cells. They play a crucial role in intercellular communication by transferring bioactive molecules such as microRNAs (miR- NAs) between cells. Recent studies have highlighted the involvement of exosome miRNAs in leukemia progression, therapy resistance, and disease prognosis. Understanding the mechanisms underlying exosome-mediated miRNA transfer in leukemia may provide valuable insights into disease pathogenesis and potential therapeutic targets. Furthermore, exosomes can serve as non-invasive biomarkers for leukemia diagnosis and monitoring treatment response. Studies have demonstrated that high levels of galectin-3 are associated with worse clinical outcomes in patients with leukemia. Understanding the specific mechanisms by which galectin-3 contributes to leukemogenesis could provide valuable insights into novel therapeutic strategies for this devastating disease. We characterized the abundance of exosomes as a function of the number and distribution in the samples. 48 h post downregulation of the LGALS3 gene, we observed a 1.2-fold increase in the release of exosomes compared to the control.
Keywords
Leukemia, Downregulation, Exosomes, Extracellular vesicles
Introduction
In recent years, the field of cancer research has witnessed remarkable advancements that have revolutionized our understanding and treatment of the disease. Among the many breakthroughs, one area of intense focus has been the study of exosomes and their role in leukemia microRNA (Amin AH, et al., 2022; Araújo HV, et al., 2022; Aucher A, et al., 2013). This cutting-edge research has the potential to not only enhance our understanding of leukemia, but also pave the way for novel diagnostic and therapeutic strategies (Barrera-Ramirez J, et al., 2017). Exosomes are small, membrane-bound vesicles that are released by various cell types, including cancer cells. They are known to carry a cargo of various molecules, including microRNAs-tiny strands of RNA that regulate gene expression. Scientists have long believed that these exosomes and their microRNA cargo play a crucial role in intercellular communication, facilitating the exchange of information between cells, both in healthy and disease states (Araújo HV, et al., 2022; Barrera-Ramirez J, et al., 2017; Bi J, et al., 2021; Bum-Erdene K, et al., 2022). In the context of leukemia, understanding the role of exosome microRNAs has proven to be of utmost importance. Leukemia is a complex and heterogeneous group of blood cancers that arise from the abnormal growth and proliferation of white blood cells. Traditional diagnostic and treatment methods have often fallen short in effectively targeting leukemia cells due to the subtle intricacies of the disease. MicroRNAs (mRNAs), on the other hand, have emerged as powerful regulators of gene expression, with the ability to fine-tune the activity of key oncogenes and tumor suppressor genes. By studying the specific microRNAs carried by exosomes in patients with leukemia, researchers have begun to uncover important clues about the underlying mechanisms that lead the disease (Chen X, et al., 2022; Di Pace AL, et al., 2023; Fang Z, et al., 2020; Feng J, et al., 2023). These microRNAs hold tremendous potential as diagnostic markers, allowing for early detection and personalized treatment strategies. In recent years, advancements in microfluidic technology have provided researchers with a powerful tool for the isolation and analysis of exosomes. Microfluidic devices allow for the precise manipulation of minute volumes of fluids, enabling the efficient capture and characterization of exosomes from patient samples. This technology has significantly accelerated our understanding of exosome-mediated communication in leukemia, thereby elucidating the role of microRNAs in the disease progression (Jiang L, et al., 2018; Li C, et al., 2022; Li H, et al., 2022). To date, several drugs and therapeutic approaches have been taken to address hematological malignancies including leukemia. However, there is still no efficient therapy for different types of leukemia. Finding a potential biomarker for leukemia diagnosis may lead to targeted therapy and finally a more favorable prognosis or treatment perspective in patients with leukemia. Recently, the role of the galectin (LGALS) family, carbohydrate-binding proteins, has been highlighted in many cancers including breast cancer, melanoma, lung cancer, sarcoma, gastric cancer, and leukemia. Galectin-3 (LGALS3) is a member of the LGALS family involving numerous biological processes such as cell adhesion, cell activation and chemoattraction, cell growth and differentiation, cell cycle, and apoptosis (Gao N, et al., 2019; Gao N, et al., 2017; Giordano M, et al., 2013; Hu K, et al., 2015). LGALS3 also interacts with several partners in the Extracellular Matrix (ECM) and thus plays a key regulatory role in maintaining the Tumor Microenvironment (TME) and cancer progression. LGALS3 was suggested to be involved in tumor-promoting mechanisms via interaction with BCL2, RAS, and beta-catenin. Previous studies revealed a high level of serum galectin in lymphoblastic leukemia compared to normal controls. Later, studies also supported the association between poor prognosis and elevated LGALS3 gene expression in patients with Acute Myeloblastic Leukemia (AML). Further studies revealed less adherent property for LGALS3 proteins to AML-derived cells after LGALS3 gene silencing and consequently sensitizing the tumor cells to chemotherapy (Li C, et al., 2022; Lepur A, et al., 2012; Liu W, et al., 2020). The mechanism of LGALS3 molecular function involved in leukemia survival has not yet been clarified. Several studies reported the LGALS3 regulation of tumor progression via miRNAs. miRNAs are small noncoding RNA, which bind to mRNAs and inhibit protein expression. Therefore, miRNA acts in wide range of cellar regulatory processes such as gene expression, epigenetic regulation, immune regulation, hematopoietic function modulation. Finding a miRNA biomarker might assist the early diagnosis and provide a therapy guide concerning recurrent Acute Lymphocytic Leukemia (ALL), which is the most leading cause of death in ALL patients. Many miRNAs are released to extracellular matrix by exosomes and interact with TEM. miRNA role is controversial. Since they act as an oncogene or a tumor suppressor they can be associated with metastasis, escape from immune system, epigenetic regulation, and hematopoietic function. Many ALL deaths are due to the lack of robust biomarkers for early detection and assistance in efficient therapy. Exosomes are newly revealed to reflect the health status; hence are a favorable biomarker to distinguish various cancers. Exosomes are 30-150 nm Extracellular Vesicles (EVs), derived from endosome and released by many types of cells into the ECM. They are detectable in body fluids including blood, urine, CSF and saliva (Mardani R, et al., 2019; Nakayama R, et al., 2014; Pena C, et al., 2014). Exosomes are responsible for cell-cell communication by harboring cellular cargoes containing proteins, RNAs, lipids and metabolites. Here we investigate the impact of LGALS3 gene silencing in miRNA profiling of large exosomes in JKB1 cells, derived from an ALL-cell line in order to find a potential biomarker for the diagnosis and treatment of ALL.
Material and Methods
Cell culture
JKB1 cells were purchased from Pasteur Institute (Tehran, Iran) and cultured in RPMI-1640 medium supplemented with 15% Phosphate Buffered Saline (FBS), 1.5% L-glutamine and 1% penicillin/streptomycin and then incubated in CO2 5% and 37°C. The medium was replaced by each 24 hours.
Basal cell proliferation assay
Methyl-Thiazole-Tetrazolium (MTT) (Sigma-Aldrich, London, UK) assay was done according to the standard protocol, briefly, cells were seeded 2 × 104 cells per well. Then 10 μl MTT solution, 5 mg/ml, was added to each well to the final concentration of 0.5 mg/ml and the cells were incubated in 37°C, 5% CO2 for 72 hours. After the generation of purple crystals, 100 μl Dimethyl Sulfoxide (DMSO) (Sigma-Aldrich, London, UK) was added to each well and they were incubated for another 4 hours in the dark. The Optical Density (OD) was finally measured at 570 nm by an Enzyme-Linked Immunosorbent Assay (ELISA) reader (BioTek, CA, USA). Means and standard deviations were calculated using 3 independent experiments and cell proliferation graph was illustrated using Microsoft Excel software.
LGALS3 silencing
The silencing of LGALS3 gene was performed as described previously. Briefly, 3 different were designed by Invitrogen’s BLOCK-iT™ RNAi Designer and synthesized by the Metabion, Germany. They were then transfected with gal-3 or Negative Control (NC) siRNA for 1 day according to the supplier’s protocol (sc-35443, Santa Cruz Biotechnology, Santa Cruz, CA, USA) in the presence of 25 ng/ml M-CSF. The end of transfection was regarded as time zero.
Western blotting
The western blot analysis was employed to confirm the LGALS3 silencing in lysed cells. In short, JKB1 cells were lysed using Radioimmunoprecipitation Assay (RIPA) buffer containing protease inhibitors, and then the total protein was extracted and protein concentration was assessed using the BCA kit. The Sodium Dodecyl-Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE) was then applied for protein separation using 10% polyacrylamide gel. The protein bands then were transferred to a Poly Vinylidene Fluoride (PVDF) membrane and the membrane was blocked using skim milk, 5%, at Room Temperature (RT), overnight. Anti-GALS-3 (1:300) and anti-β-actin (1:5000) (Santa Cruz Biotechnology, Dallas, USA) were incubated, at 4°C, for 1 hour. Next, the secondary antibody, Rabbit anti-mouse antibody-HRP conjugated, (1:5000) was added at RT, for 1 hour. The protein bands visualized using by Immobilon Western Chemiluminescent HRP Substrate detection system (Merck milli-pore, Burlington, Massachusetts, USA) according to the manufacturer’s instruction.
Large exosome separation and characterization
To isolate large exosomes, 10000 JKB1 cells cultured and centrifuged at 1000 g, for 5 minutes and then to remove debris, the Supernatants were centrifuged for another 40 minutes at 4500 g. The exosomes were then isolated using plasma/serum exosome purification kit (cat-57400, Norgen Biotechnology, Canada) according to the manufacturer’s instructions. Dynamic Light Scattering (DSL) technique was employed to evaluate the hydrodynamic size distribution of isolated exosomes using ZetaSizer (Malvern Panalytical, CA, USA). Exosomes were diluted to the concentration of 50 μg/μl and sample loaded into the cuvette after equilibration of temperature to 25°C. The DLS was run in 3 replicates, recorded for 10 s at a laser wavelength 663 nm. Then the exosomes shapes were analyzed using Scanning Electron Microscope (SEM) (LAbX, CA, USA). Primarily, exosomes were fixed with 1.5% glutaraldehyde (Sigma-Aldrich, London, UK) in PBS for 10 min, following the washing steps; they were dehydrated using ethanol solution from 40% to 98% and then incubated at room temperature, overnight to get dried. After gold sputtering (Sputter Coater 150A, Edwards, UK), exosomes were analyzed using SEM at a working voltage of 33 kV.
miRNA profiling of large exosomes
Total RNA was extracted using the microRNA Purification Kit (cat-21300, Norgen Biotechnology, Canada) according to the manufacturer’s protocol. Total RNA (20 to 50 ng) was then reverse transcribed using the TaqMan MicroRNA Reverse Transcription Kit (TaKaRa, Japan) according to the manufacturer’s instructions. The cDNA template was then applied in microRNA profiling of exosomes using specific microRNA primers according to previously published and run on an CFX384 Touch Real-Time PCR Detection System (Bio-Rad, CA, USA) (Jiang L, et al., 2018, Jiang D, et al., 2022; Lin X, et al., 2020; Peng D, et al., 2018; Rzepiel A, et al., 2023; Shi XF, et al., 2017). Each miRNA was assayed in triplicate on by Real-Time Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR). Data were analyzed by the 2-∆∆Ct method. Relative expression levels of each miRNA were normalized using reference of normal lymphocyte and RNU6A gene as an endogenous control.Statistical analysis
Comparisons between experimental groups were performed using a two tailed, unpaired, Student’s t-test. Pearson’s Chi-Square (χ2) test was used to assess the correlation between fold-change of target miRNA in silenced cells and silenced free JKB1 cells.
Results and Discussion
RNA interference in JKB1 cells post transcriptionally downregulates LGALS3 expression
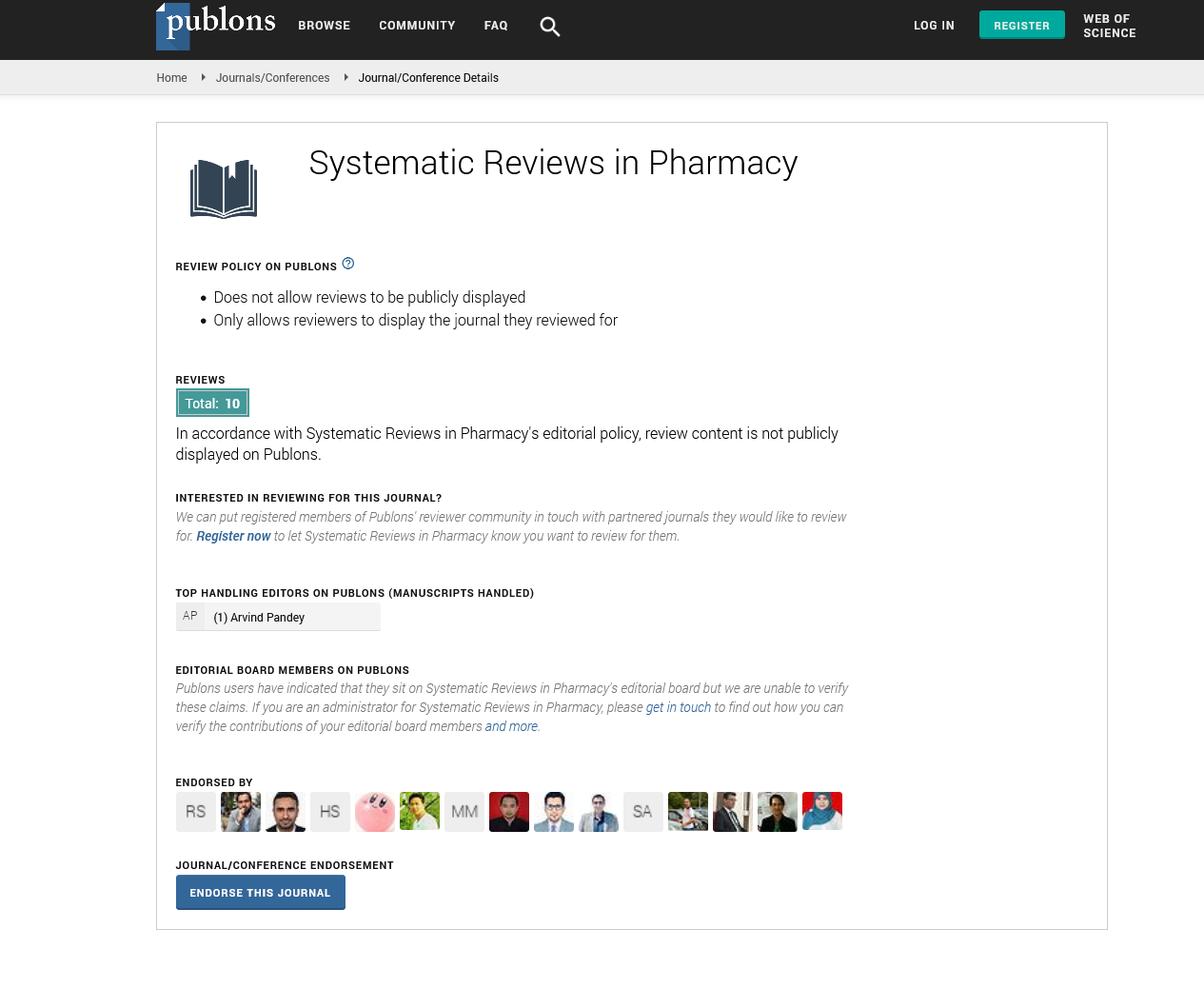
Four human LGALS3 siRNA, were tested for reducing LGALS3 expression in human JKB1 cell lines. The most efficient siRNA was retained for the experiment. The pGB vector was cut with BamH 1 and Xba 1 restriction enzymes. The LGALS3 targeting cassette construct was inserted downstream of the PhU6 promoter to form the pGB-siGALS3 plasmid. Human JKB1 cells were transfected with the pGB-siGALS3 plasmid (Figure 1). The targeting specificity was also confirmed by employing qRT-PCR and immunoblotting. Analysis of the gene expression depicted a 2.9-fold decrease of the constitutive LGALS3 expression from 3.09 to 0.18 in JKB1 cells silenced with the pGB-siGALS3 vector (Figure 2).
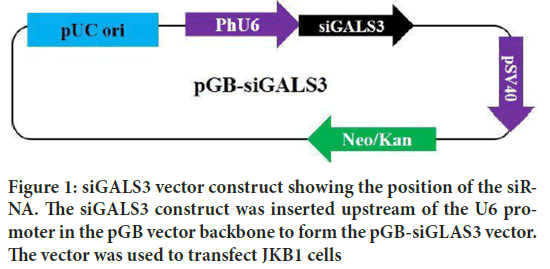
Figure 1: siGALS3 vector construct showing the position of the siRNA. The siGALS3 construct was inserted upstream of the U6 promoter in the pGB vector backbone to form the pGB-siGLAS3 vector. The vector was used to transfect JKB1 cells
Figure 2: siGALS3 downregulates the expression of LGALS3 gene in
JKB1 cells (A) LGALS3 gene expression levels were analyzed post
transfection employing qRT-PCR and the relative expression of
LGALS3 was normalized using β-Actin as the endogenous control;
(B) Immunoblot quantification demonstrating the knockdown of
LGALS3 gene, β-Actin as the loading control
Note: ****p<0.0001, all data are represented in triplicates as mean
± SD
The growth of JKB1 cells in vitro is influenced by LGALS3 gene
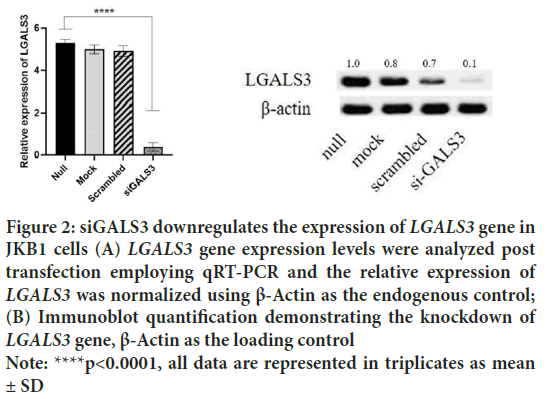
JKB1 cells were cultured in RPMI media for a total of eight days and daily checks were performed to evaluate the oncogenic effect of LGALS3 gene on the cultured cells. The initially seeded cells were passaged after three days and newly seeded cell were monitored closely to determine any eventual impact of our experimental intervention on the cells. Cell proliferation assay was performed 12 hours post seeding and every 24 hours thereafter. The cells reached an exponential phase by 72 hours post incubation in all four experimental setups and changes in the proliferation state became prominent in the days after transfection at 72 hours in the siGALS3 setup. The null cell line setup reached maximum proliferation at 120 hours post incubation and started declining after 144 hours. By 192 hours, all cells in the null setup were dead. Contrarily, the siGALS3 setup reached an early death phase by 144 hours (Figure 3). This early death experience could be attributed to the effect of silencing of the LGALS3 gene in this setup, as the other setups had similar growth profile as the control setup (null cell line).
Figure 3: Scanning electron micrograph illustrating the presence of
large exosomes in JKB1 cell supernatant
Note:  siGALS3
siGALS3
Large exosomes are uniformly and fairly distributed across JKB1 supernatants
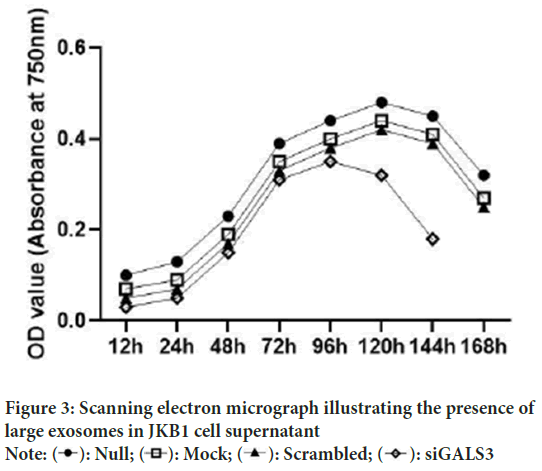
To ascertain the occurrence of large exosomes in extracted supernatants from the JKB1 cultured cells, stubs were coated with gold to improve and facilitate their detection. Initially, Scanning Emission Microscope (SEM) was used to designate the molecules down to an observable imaging resolution proceeded by transmission electron microscopic observation. These two methods could provide the required information to distinguish the shape and denote the relative abundance of the observed particles within the supernatant. The supernatants collected from both the siRNA control setup and the siGALS3 (test) revealed the presence of exosomes at high concentrations. Visualization and biochemical characterization depicted a uniform distribution of the exosomes in a state relative to their native conformation (Figure 4).
Figure 4: Scanning electron micrograph showing the presence of
large exosomes in the JKB1 cell supernatant
Note:  Fixation and staining with gold material followed by
cryo-SEM shows the abundance of large exosomes with respect to
their native state.
Fixation and staining with gold material followed by
cryo-SEM shows the abundance of large exosomes with respect to
their native state.
Downregulation of LGALS3 gene precipitates the secretion of large exosomes
The isolation of large exosomes was executed utilizing the Norgen-Biotek exosome isolation kit protocol. Antibodies against CD63, CD326, and TBP (Tim4-binding phosphatidylserine) exosome surface markers were utilized to ascertain the presence of exosomes in the supernatant of test and control cell lines. Quantification by employing the DSL depicted semi-uniformity by serial dilution. Samples were analyzed at different time points commencing at 12 hours post transfection to 48 hours post transfection. Initial counts following serial dilution revealed fairly-uniform sized particles at 0.01 ml up to 0.1 ml particle counts. At 1 ml counts 48 hours post-transfection when LGALS3 was completely downregulated, quantification depicted a high abundance of exosomes in the silenced cell supernatant than in the non-silenced control setup. We characterized the abundance of exosomes as a function of the number and distribution in the samples. 48 hours post downregulation of the LGALS3 gene, we observed a 1.2-fold increase in the release of exosomes compared to the control.
Hematologic Malignancies (HMs) represent a heterogeneous group of hematopoietic neoplasms commonly characterized by the abnormal production of blood cells. Recent studies revealed that these biological macromolecules can function in intercellular communication through exosome trafficking from immune cells, fibroblast and cancer cells to a range of different recipient cells (Yang Y, et al., 2021; Yuan Y, et al., 2023; Zhang F, et al., 2020). Exosome miRNAs do not repress target gene expression or activate the endosomal TLR8 receptor in recipient cells. Thus, exosome-mediated cell-cell communications could play potential important roles during leukemia development. exosomes that are released from tumor cells, which are involved in tumor angiogenesis, can be taken up by vascular endothelial cells, thereby encouraging angiogenesis (Wang JJ, et al., 2015; Yan W, et al., 2021; Zhao C, et al., 2019; Zhao C, et al., 2022). By transferring molecules from one cell to another, exosomes from certain cells of the immune system, such as dendritic cells and B cells, may play a functional role in mediating adaptive immune responses to pathogens and tumors. In recent years, exosomes have received more attention as a promising tumor screening, diagnostic and prognostic biomarker. By binding to the 3’ Untranslated Regions (3UTR) of the target mRNA, the small RNA molecules regulate gene expression primarily at the post-translational levels (Tarighat SS, et al., 2021; Tutar Y, 2014; Wang D, et al., 2021; Wang J, et al., 2022; Yamamoto-Sugitani M, et al., 2011). Thus, it can be said that exosomes derived from leukemic cells in AML have the potential to modulate the immune system and may influence disease progression as well as response to therapy (Shi XF, et al., 2017). Exosomes are found in most, if not all, biological fluids including urine, blood, ascites and cerebrospinal fluid. In brief, Virus-Like Particles (VLPs) are thermoresistant and more stable than lipid-based particles, which protect miRNAs from rapid degradation by nucleases (Quirico L and Orso F, 2020; Rios de los Rios J, et al., 2022; Ruvolo PP, 2016; Turi M, et al., 2023). Accordingly, we can conclude that exosome miRNAs can provide information on the molecular characteristics of the cells by which they were secreted and be endowed with the capacity to reprogram distant target cells. Extracellular Vehicles (EVs), including exosomes, micro vesicles and apoptotic bodies, participate in intercellular communication, and particularly, in paracrine and endocrine signaling. Recently, it has been shown that exosomes secreted from Mesenchymal Stem Cells (MSCs) play a critical role in MSC-mediated paracrine effects via transfer of miRNAs (Shi XF, et al., 2017; Paz H, et al., 2018; Vinnai JR, et al., 2017; Wallace JA and O’Connell RM, 2017). Additionally, exosomes function mainly by influencing gene expression and signaling pathways, which are closely related to miRNA regulation of cellular communication. Certain miRNA profiles have also been shown to associate specifically with pediatric ALL and reveal distinct expression profiles based on the cell of origin and cytogenetic subtype. Overall, delivery of AAV-mediated miRNAs holds high importance in cancer therapy (Aucher A, et al., 2013; Barrera-Ramirez J, et al., 2017; Fei F, et al., 2015; Otmani K, et al., 2023). New advances in gene therapy, such as potential delivery vehicles for miRNAs, could facilitate development of anti-miR-1246 therapeutic strategies. Clarifying the role of tumor-derived exosomes in cancer progression may change aspects of cancer treatment, leading to personalized medicine. miRNAs transfer can also cause the physiological changes in their recipient cells, which was demonstrated by miRNAs moving from cancer cells to endothelial cells, promoting tumor metastasis. Researchers obtained exosomes from self-derived dendritic cells and then decorated exosomes to express membrane protein Lamp2b and a neuron-specific RVG peptide in order to deliver cargoes specifically to recipient neural cells. Regarding new separation methods, immuno-affinity purification and microfluidics-based isolation both have high efficiency and good purification (Bi J, et al., 2021; Asgarian-Omran H, et al., 2010; Boyiadzis M and Whiteside TL, 2016). Here, we summarize the roles played by different miRNAs in childhood leukemia, focusing primarily on their use as diagnostic tools and potential therapeutic targets, as well as a role in predicting treatment outcome. The diagnostic and prognostic significance of Galectin-3 in leukemia patients is a topic of growing interest in the field of hematology. Galectin-3, a multifunctional carbohydrate-binding protein, has been found to play a crucial role in various cellular processes, including cell adhesion, proliferation, and apoptosis. In the context of leukemia, studies have shown that galectin-3 expression levels are often dysregulated and can serve as potential diagnostic markers for different subtypes of the disease (Bum-Erdene K, et al., 2022; Asgarian-Omran H, et al., 2010; Cheng YL, et al., 2011; Fei F, et al., 2022). Galectin-3 has been implicated in the prognosis of leukemia patients. High levels of galectin-3 expression have been associated with poor clinical outcomes and increased resistance to chemotherapy. Conversely, lower galectin-3 expression levels have been linked to better treatment response and improved survival rates. Understanding the diagnostic and prognostic significance of galectin-3 in leukemia patients holds immense potential for personalized medicine approaches. Inhibition of galectin-3 expression or activity: Developing small molecules or antibodies that specifically target galectin-3 and inhibit its expression or activity could be a successful therapeutic strategy. This approach aims to disrupt the interaction between galectin-3 and leukemia cells, thereby inhibiting cell proliferation, migration, and survival. Combination therapy with existing treatments: Combining galectin-3 inhibitors with conventional chemotherapy drugs or targeted therapies could enhance their efficacy and overcome drug resistance in leukemia. Due to their great potential, exosome miRNAs can be used as an excellent non-invasive tool for early diagnosis, prognosis and prediction of treatment success or drug resistances in this pathology. The many of studies demonstration that the new method is also appropriate for analysis of the miRNA content of large exosomes, and that >60% of the miRNAs are expressed at comparable levels between donor cells and derivative exosomes, proposes that some miRNAs are travelled into the particles because of subcellular position, indicating that the miRNA content of the exosome is illustrative of the donor cell manifestation. Consequently, large exosomes are a potential source of circulating markers of leukemia response to treatment. Notably, GAL-3 regulator miRNA hsa-miR-23b and hsa-miR-20a-5p were identified in extra cellular vesicles. In addition, some of the miRNAs expressed at higher levels in EVs from the silenced cell line, hsa-mir-424, was recently incorporated into a tissue miRNA signature of clinical progression in patients with cervical cancer. Interestingly, pathway analysis demonstrated that the top 2 biological functions affected by the 2 miRNAs enriched in exosome from the leukemogenic cells are usually altered in leukemia (Jiang D, et al., 2022; Beeraka NM, et al., 2020; Damanti CC, et al., 2021; Hornick NI, et al., 2015).
Conclusion
The miRNAs expressed at comparable levels, our data also show that several miRNAs are differentially expressed in contributor cells and exosomes, data has not shown, suggesting that the boxing of miRNAs into exosomes does occur in portion by dynamic selection. Although differences in miRNA levels in exosomes and donor cells have been reported for other types of cell lines, this is the first localized comparison of the miRNA profile of exosomes derived from silenced and non-silenced JKB1 as a leukemia cell model. In line with this hypothesis, our functional studies demonstrated that hsa-miR-23b, one of the top 10 expressed miRNAs in exosomes from the silenced JKB1, could produce biological effects in receptor biosynthetic process and impress to cell to cell talking.
References
- Amin AH, Al Sharifi LM, Kakhharov AJ, Opulencia MJ, Alsaikhan F, Bokov DO, et al. Role of Acute Myeloid Leukemia (AML)-derived exosomes in tumor progression and survival. Biomed Pharmacother. 2022; 150: 113009.
[Crossref] [Google Scholar] [Pubmed]
- Araújo HV, Sakamoto LH, Bacal NS, Epelman S, Real JM. MicroRNAs and exosomes: Promising new biomarkers in acute myeloid leukemias? Einstein (Sao Paulo). 2022; 20: eRB5954.
[Crossref] [Google Scholar] [Pubmed]
- Aucher A, Rudnicka D, Davis DM. MicroRNAs transfer from human macrophages to hepato-carcinoma cells and inhibit proliferation. J Immunol. 2013; 191(12): 6250-6260.
[Crossref] [Google Scholar] [Pubmed]
- Barrera-Ramirez J, Lavoie JR, Maganti HB, Stanford WL, Ito C, Sabloff M, et al. Micro-RNA profiling of exosomes from marrow-derived mesenchymal stromal cells in patients with acute myeloid leukemia: Implications in leukemogenesis. Stem Cell Rev Rep. 2017;13: 817-825.
[Crossref] [Google Scholar] [Pubmed]
- Bi J, Pu Y, Yu X. Exosomal circ_0004136 enhances the progression of pediatric acute myeloid leukemia depending on the regulation of miR-570-3p/TSPAN3 axis. Anticancer Drugs. 2021; 32(8): 802-811.
[Crossref] [Google Scholar] [Pubmed]
- Bum-Erdene K, Collins PM, Hugo MW, Tarighat SS, Fei F, Kishor C, et al. Novel selective galectin-3 antagonists are cytotoxic to acute lymphoblastic leukemia. J Med Chem. 2022; 65(8): 5975-5989.
[Crossref] [Google Scholar] [Pubmed]
- Chen X, Chen Y, Zhang M, Cheng H, Mai H, Yi M, et al. HucMSC exosomes promoted imatinib-induced apoptosis in K562-R cells via a miR-145a-5p/USP6/GLS1 axis. Cell Death Dis. 2022; 13(1): 92.
[Crossref] [Google Scholar] [Pubmed]
- Di Pace AL, Pelosi A, Fiore PF, Tumino N, Besi F, Quatrini L, et al. MicroRNA analysis of natural killer cell-derived exosomes: The microRNA let-7b-5p is enriched in exosomes and participates in their anti-tumor effects against pancreatic cancer cells. OncoImmunology. 2023; 12(1): 2221081.
[Crossref] [Google Scholar] [Pubmed]
- Fang Z, Wang X, Wu J, Xiao R, Liu J. High serum extracellular vesicle miR-10b expression predicts poor prognosis in patients with acute myeloid leukemia. Cancer Biomark. 2020; 27(1): 1-9.
[Crossref] [Google Scholar] [Pubmed]
- Feng J, Wang Y, Li B, Yu X, Lei L, Wu J, et al. Loss of bisecting GlcNAcylation on MCAM of bone marrow stoma determined pro-tumoral niche in MDS/AML. Leukemia. 2023; 37(1): 113-121.
[Crossref] [Google Scholar] [Pubmed]
- Jiang L, Deng T, Wang D, Xiao Y. Elevated serum exosomal miR-125b level as a potential marker for poor prognosis in intermediate-risk acute myeloid leukemia. Acta Haematol. 2018; 140(3): 183-192.
[Crossref] [Google Scholar] [Pubmed]
- Li C, Long X, Liang P, Liu Z, Wang C, Hu R. Analysis of microRNA expression profiles in exosomes derived from acute myeloid leukemia by p62 knockdown and effect on angiogenesis. PeerJ. 2022; 10: e13498.
[Crossref] [Google Scholar] [Pubmed]
- Li H, Xie C, Lu Y, Chang K, Guan F, Li X. Exosomal miR92a promotes cytarabine resistance in myelodysplastic syndromes by activating Wnt/β-catenin signal pathway. Biomolecules. 2022; 12(10): 1448.
[Crossref] [Google Scholar] [Pubmed]
- Gao N, Wang XX, Sun JR, Duan PF, Liu ZY, Liu RT, et al. Galectin-3 expression and its clinical significance in acute myeloid leukemia with positive AML1/ETO fusion gene. Sichuan Da Xue Xue Bao Yi Xue Ban. 2019; 50(1): 109-14.
[Google Scholar] [Pubmed]
- Gao N, Yu WZ, Guo NJ, Wang XX, Sun JR. Clinical significance of galectin-3 in patients with adult acute myeloid leukemia: A retrospective cohort study with long-term follow-up and formulation of risk scoring system. Leuk Lymphoma. 2017; 58(6): 1394-1402.
[Crossref] [Google Scholar] [Pubmed]
- Giordano M, Croci DO, Rabinovich GA. Galectins in hematological malignancies. Curr Opin Hematol. 2013; 20(4): 327-335.
[Crossref] [Google Scholar] [Pubmed]
- Hu K, Gu Y, Lou L, Liu L, Hu Y, Wang B, et al. Galectin-3 mediates bone marrow microenvironment-induced drug resistance in acute leukemia cells via Wnt/β-catenin signaling pathway. J Hematol Oncol. 2015; 8(1): 1.
[Crossref] [Google Scholar] [Pubmed]
- Lepur A, Carlsson MC, Novak R, Dumić J, Nilsson UJ, Leffler H. Galectin-3 endocytosis by carbohydrate independent and dependent pathways in different macrophage like cell types. Biochim Biophys Acta. 2012; 1820(7): 804-818.
[Crossref] [Google Scholar] [Pubmed]
- Liu W, Xiao K, Ren L, Sui Y, Chen J, Zhang T, et al. Leukemia cells apoptosis by a newly discovered heterogeneous polysaccharide from Angelica sinensis (Oliv.) Diels. Carbohydr Polym. 2020; 241: 116279.
[Crossref] [Google Scholar] [Pubmed]
- Mardani R, Abadi MH, Motieian M, Taghizadeh‐Boroujeni S, Bayat A, Farsinezhad A, et al. MicroRNA in leukemia: Tumor suppressors and oncogenes with prognostic potential. J Cell Physiol. 2019; 234(6): 8465-8486.
[Crossref] [Google Scholar] [Pubmed]
- Nakayama R, Kuroda J, Taniyama N, Yamamoto-Sugitani M, Wada S, Kiyota M, et al. Suppression of SERPINA1-albumin complex formation by galectin-3 overexpression leads to paracrine growth promotion of chronic myelogenous leukemia cells. Leuk Res. 2014; 38(1): 103-108.
[Crossref] [Google Scholar] [Pubmed]
- Pena C, Mirandola L, Figueroa JA, Hosiriluck N, Suvorava N, Trotter K, et al. Galectins as therapeutic targets for hematological malignancies: A hopeful sweetness. Ann Transl Med. 2014; 2(9): 87.
[Crossref] [Google Scholar] [Pubmed]
- Jiang D, Wu X, Sun X, Tan W, Dai X, Xie Y, et al. Bone mesenchymal stem cell-derived exosomal microRNA-7-5p inhibits progression of acute myeloid leukemia by targeting OSBPL11. J Nanobiotechnology. 2022; 20(1): 1-9.
[Crossref] [Google Scholar] [Pubmed]
- Lin X, Ling Q, Lv Y, Ye W, Huang J, Li X, et al. Plasma exosome-derived microRNA-532 as a novel predictor for acute myeloid leukemia. Cancer Biomark. 2020; 28(2): 151-158.
[Crossref] [Google Scholar] [Pubmed]
- Peng D, Wang H, Li L, Ma X, Chen Y, Zhou H, et al. miR-34c-5p promotes eradication of acute myeloid leukemia stem cells by inducing senescence through selective RAB27B targeting to inhibit exosome shedding. Leukemia. 2018; 32(5): 1180-1188.
[Crossref] [Google Scholar] [Pubmed]
- Rzepiel A, Horváth A, Kutszegi N, Gézsi A, Sági JC, Almási L, et al. MiR-128-3p as blood based liquid biopsy biomarker in childhood acute lymphoblastic leukemia. Mol Cell Probes. 2023; 67: 101893.
[Crossref] [Google Scholar] [Pubmed]
- Shi XF, Wang H, Kong FX, Xu QQ, Xiao FJ, Yang YF, et al. Exosomal miR-486 regulates hypoxia-induced erythroid differentiation of erythroleukemia cells through targeting Sirt1. Exp Cell Res. 2017; 351(1): 74-81.
[Crossref] [Google Scholar] [Pubmed]
- Yang Y, He H, He J, Gu X, Hu P, Zuo R, et al. Hyperleukocytic acute leukemia circulating exosomes regulate HSCs and BM-MSCs. J Healthc Eng. 2021; 2021.
[Crossref] [Google Scholar] [Pubmed]
- Yuan Y, Tan S, Wang H, Zhu J, Li J, Zhang P, et al. Mesenchymal stem cell-derived exosomal miRNA-222-3p increases Th1/Th2 ratio and promotes apoptosis of acute myeloid leukemia cells. Anal Cell Pathol. 2023; 2023.
[Crossref] [Google Scholar] [Pubmed]
- Zhang F, Lu Y, Wang M, Zhu J, Li J, Zhang P, et al. Exosomes derived from human bone marrow mesenchymal stem cells transfer miR-222-3p to suppress acute myeloid leukemia cell proliferation by targeting IRF2/INPP4B. Mol Cell Probes. 2020; 51: 101513.
[Crossref] [Google Scholar] [Pubmed]
- Wang JJ, Wang ZY, Chen R, Xiong J, Yao YL, Wu JH, et al. Macrophage-secreted exosomes delivering miRNA-21 inhibitor can regulate BGC-823 cell proliferation. Asian Pac J Cancer Prev. 2015; 16(10): 4203-4209.
[Crossref] [Google Scholar] [Pubmed]
- Yan W, Song L, Wang H, Yang W, Hu L, Yang Y. Extracellular vesicles carrying miRNA-181b-5p affects the malignant progression of acute lymphoblastic leukemia. J Transl Med. 2021; 19(1): 1-9.
[Crossref] [Google Scholar] [Pubmed]
- Zhao C, Du F, Zhao Y, Wang S, Qi L. Acute myeloid leukemia cells secrete microRNA-4532-containing exosomes to mediate normal hematopoiesis in hematopoietic stem cells by activating the LDOC1-dependent STAT3 signaling pathway. Stem Cell Res Ther. 2019; 10(1): 1-2.
[Crossref] [Google Scholar] [Pubmed]
- Zhao C, Zhao Y, Zhao J, Meng G, Huang S, Liu Y, et al. Acute myeloid leukemia cell-derived extracellular vesicles carrying microRNA-548ac regulate hematopoietic function via the TRIM28/STAT3 pathway. Cancer Gene Ther. 2022; 29(7): 918-929.
[Crossref] [Google Scholar] [Pubmed]
- Tarighat SS, Fei F, Joo EJ, Abdel-Azim H, Yang L, Geng H, et al. Overcoming microenvironment-mediated chemoprotection through stromal galectin-3 inhibition in acute lymphoblastic leukemia. Int J Mol Sci. 2021; 22(22): 12167.
[Crossref] [Google Scholar] [Pubmed]
- Tutar Y. miRNA and cancer; computational and experimental approaches. Curr Pharm Biotechnol. 2014; 15(5): 429.
[Crossref] [Google Scholar] [Pubmed]
- Wang D, Ming X, Xu J, Xiao Y. Circ_0009910 shuttled by exosomes regulates proliferation, cell cycle and apoptosis of acute myeloid leukemia cells by regulating miR‐5195‐3p/GRB10 axis. Hematol Oncol. 2021; 39(3): 390-400.
[Crossref] [Google Scholar] [Pubmed]
- Wang J, Gao N, Wang X, Yu W, Li A. Prognostic factors in acute myeloid leukemia with t (8; 21)/AML1-ETO: Strategies to define high-risk patients. Indian J Hematol Blood Transfus. 2022; 38(4): 631-637.
[Crossref] [Google Scholar] [Pubmed]
- Yamamoto-Sugitani M, Kuroda J, Ashihara E, Nagoshi H, Kobayashi T, Matsumoto Y, et al. Galectin-3 (Gal-3) induced by leukemia microenvironment promotes drug resistance and bone marrow lodgment in chronic myelogenous leukemia. Proc Natl Acad Sci U S A. 2011; 108(42): 17468-17473.
[Crossref] [Google Scholar] [Pubmed]
- Quirico L, Orso F. The power of microRNAs as diagnostic and prognostic biomarkers in liquid biopsies. Cancer Drug Resist. 2020; 3(2): 117.
[Crossref] [Google Scholar] [Pubmed]
- Rios de los Rios J, Enciso J, Vilchis-Ordoñez A, Vázquez-Ramírez R, Ramirez-Ramirez D, Balandrán JC, et al. Acute lymphoblastic leukemia-secreted miRNAs induce a proinflammatory microenvironment and promote the activation of hematopoietic progenitors. J Leukoc Biol. 2022; 112(1): 31-45.
[Crossref] [Google Scholar] [Pubmed]
- Ruvolo PP. Galectin 3 as a guardian of the tumor microenvironment. Biochim Biophys Acta. 2016; 1863(3): 427-437.
[Crossref] [Google Scholar] [Pubmed]
- Turi M, Sithara A, Hofmanová L, Žihala D, Radhakrishnan D, Vdovin A, et al. Transcriptome analysis of diffuse large B-cell lymphoma cells inducibly expressing MyD88 L265P mutation identifies upregulated CD44, LGALS3, NFKBIZ, and BATF as downstream targets of oncogenic NF-κB signaling. Int J Mol Sci. 2023; 24(6): 5623.
[Crossref] [Google Scholar] [Pubmed]
- Paz H, Joo EJ, Chou CH, Fei F, Mayo KH, Abdel-Azim H, et al. Treatment of B-cell precursor acute lymphoblastic leukemia with the galectin-1 inhibitor PTX008. J Exp Clin Cancer Res. 2018; 37(1): 1-5.
[Crossref] [Google Scholar] [Pubmed]
- Vinnai JR, Cumming RC, Thompson GJ, Timoshenko AV. The association between oxidative stress-induced galectins and differentiation of human promyelocytic HL-60 cells. Exp Cell Res. 2017; 355(2): 113-123.
[Crossref] [Google Scholar] [Pubmed]
- Wallace JA, O’Connell RM. MicroRNAs and acute myeloid leukemia: Therapeutic implications and emerging concepts. Blood. 2017; 130(11): 1290-1301.
[Crossref] [Google Scholar] [Pubmed]
- Fei F, Joo EJ, Tarighat SS, Schiffer I, Paz H, Fabbri M, et al. B-cell precursor acute lymphoblastic leukemia and stromal cells communicate through galectin-3. Oncotarget. 2015; 6(13): 11378.
[Crossref] [Google Scholar] [Pubmed]
- Otmani K, Rouas R, Lagneaux L, Krayem M, Duvillier H, Berehab M, et al. Acute myeloid leukemia-derived exosomes deliver miR-24-3p to hinder the T-cell immune response through DENN/MADD targeting in the NF-κB signaling pathways. Cell Commun Signal. 2023; 21(1): 253.
[Crossref] [Google Scholar] [Pubmed]
- Asgarian-Omran H, Forghani P, Hojjat-Farsangi M, Roohi A, Sharifian RA, Razavi SM, et al. Expression profile of galectin-1 and galectin-3 molecules in different subtypes of chronic lymphocytic leukemia. Cancer Invest. 2010; 28(7): 717-725.
[Crossref] [Google Scholar] [Pubmed]
- Boyiadzis M, Whiteside TL. Plasma-derived exosomes in acute myeloid leukemia for detection of minimal residual disease: Are we ready? Expert Rev Mol Diagn. 2016; 16(6): 623-629.
[Crossref] [Google Scholar] [Pubmed]
- Cheng YL, Huang WC, Chen CL, Tsai CC, Wang CY, Chiu WH, et al. Increased galectin-3 facilitates leukemia cell survival from apoptotic stimuli. Biochem Biophys Res Commun. 2011; 412(2): 334-340.
[Crossref] [Google Scholar] [Pubmed]
- Fei F, Zhang M, Tarighat SS, Joo EJ, Yang L, Heisterkamp N. Galectin-1 and galectin-3 in B-cell precursor acute lymphoblastic leukemia. Int J Mol Sci. 2022; 23(22): 14359.
[Crossref] [Google Scholar] [Pubmed]
- Beeraka NM, Doreswamy SH, Sadhu SP, Srinivasan A, Pragada RR, Madhunapantula SV, et al. The role of exosomes in stemness and neurodegenerative diseases-chemoresistant-cancer therapeutics and phytochemicals. Int J Mol Sci. 2020; 21(18): 6818.
[Crossref] [Google Scholar] [Pubmed]
- Damanti CC, Gaffo E, Lovisa F, Garbin A, Di Battista P, Gallingani I, et al. MiR-26a-5p as a reference to normalize MicroRNA qRT-PCR levels in plasma exosomes of pediatric hematological malignancies. Cells. 2021; 10(1): 101.
[Crossref] [Google Scholar] [Pubmed]
- Hornick NI, Huan J, Doron B, Goloviznina NA, Lapidus J, Chang BH, et al. Serum exosome microRNA as a minimally-invasive early biomarker of AML. Sci Rep. 2015; 5(1): 11295.
[Crossref] [Google Scholar] [Pubmed]
Author Info
Amir Monfaredan1, Fakher Rahim2, Gholamreza Tavoosidana1, Mohammad Hossein Modarressi3, Alaviyehsadat hosseininasab4, Ali-Akbar Aghajani-Afrouzi5, Mahdi Shafiee Sabet6 and Elahe Motevaseli1*2Department of Health Sciences, Cihan university-Sulaymaniyah, Sulaymaniyah, Iraq
3Department of Medical Genetics, Tehran University of Medical Sciences, Tehran, Iran
4GeneDia Life Science Company, Tehran, Iran
5Department of Business Administration, Payame Noor University, Tehran, Iran
6Department of Neurology, School of Medicine, Tehran University of Medical Sciences, Tehran, Iran
Citation: Monfaredan A: The Downregulation of LGALS3 Gene Expression can Stimulate the Release of Exosomes in the JKB1 Cell Line
Received: 02-Nov-2023 Accepted: 16-Nov-2023 Published: 23-Nov-2023, DOI: 10.31858/0975-8453.14.11.695-700
Copyright: This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
ARTICLE TOOLS
- Dental Development between Assisted Reproductive Therapy (Art) and Natural Conceived Children: A Comparative Pilot Study Norzaiti Mohd Kenali, Naimah Hasanah Mohd Fathil, Norbasyirah Bohari, Ahmad Faisal Ismail, Roszaman Ramli SRP. 2020; 11(1): 01-06 » doi: 10.5530/srp.2020.1.01
- Psychometric properties of the World Health Organization Quality of life instrument, short form: Validity in the Vietnamese healthcare context Trung Quang Vo*, Bao Tran Thuy Tran, Ngan Thuy Nguyen, Tram ThiHuyen Nguyen, Thuy Phan Chung Tran SRP. 2020; 11(1): 14-22 » doi: 10.5530/srp.2019.1.3
- A Review of Pharmacoeconomics: the key to “Healthcare for All” Hasamnis AA, Patil SS, Shaik Imam, Narendiran K SRP. 2019; 10(1): s40-s42 » doi: 10.5530/srp.2019.1s.21
- Deuterium Depleted Water as an Adjuvant in Treatment of Cancer Anton Syroeshkin, Olga Levitskaya, Elena Uspenskaya, Tatiana Pleteneva, Daria Romaykina, Daria Ermakova SRP. 2019; 10(1): 112-117 » doi: 10.5530/srp.2019.1.19
- Dental Development between Assisted Reproductive Therapy (Art) and Natural Conceived Children: A Comparative Pilot Study Norzaiti Mohd Kenali, Naimah Hasanah Mohd Fathil, Norbasyirah Bohari, Ahmad Faisal Ismail, Roszaman Ramli SRP. 2020; 11(1): 01-06 » doi: 10.5530/srp.2020.1.01
- Manilkara zapota (L.) Royen Fruit Peel: A Phytochemical and Pharmacological Review Karle Pravin P, Dhawale Shashikant C SRP. 2019; 10(1): 11-14 » doi: 0.5530/srp.2019.1.2
- Pharmacognostic and Phytopharmacological Overview on Bombax ceiba Pankaj Haribhau Chaudhary, Mukund Ganeshrao Tawar SRP. 2019; 10(1): 20-25 » doi: 10.5530/srp.2019.1.4
- A Review of Pharmacoeconomics: the key to “Healthcare for All” Hasamnis AA, Patil SS, Shaik Imam, Narendiran K SRP. 2019; 10(1): s40-s42 » doi: 10.5530/srp.2019.1s.21
- A Prospective Review on Phyto-Pharmacological Aspects of Andrographis paniculata Govindraj Akilandeswari, Arumugam Vijaya Anand, Palanisamy Sampathkumar, Puthamohan Vinayaga Moorthi, Basavaraju Preethi SRP. 2019; 10(1): 15-19 » doi: 10.5530/srp.2019.1.3